To use this workshop
Register or log-in
choose the ‘ARTNet 2023’
Wait a minute or so
Click to open RStudio
-
In RStudio, choose ‘File’ / ‘Open File…’ / ‘vignettes/a_r.Rmd’
Introduction
This article illustrates how R can be used to understand single-cell RNAseq data, assuming that the ‘heavy lifting’ of transforming raw FASTQ files to normalized matrices of counts measuring expression of each gene in each cell has been done by, e.g., a bioinformatics core. R can be used to summarize biological sample and cell attributes, e.g., the number of donors and cells per donor. Visualizations like UMAP plots showing cell expression patterns in two-dimensional space can be easily generated and annotated. Individual genes can be annotated with additional information, e.g., with a description of the gene or of the genes in particular pathways. The next article introduces more comprehensive work flows.
Cell summary
We will use the dplyr package for data manipulation.
Read a ‘csv’ file summarizing infomration about each cell in the experiment.
## use `file.choose()` or similar for your own data sets
cell_data_csv <- system.file(package = "ARTNet2023", "scrnaseq-cell-data.csv")
cell_data <- readr::read_csv(cell_data_csv)
cell_data |>
glimpse()
#> Rows: 31,696
#> Columns: 37
#> $ cell_id <chr> "CMGpool_AAACCCAAGGACAACC", "…
#> $ UMAP_1 <dbl> -5.9931696, -6.7217583, -9.01…
#> $ UMAP_2 <dbl> -1.9311870, -1.3059803, -3.39…
#> $ donor_id <chr> "pooled [D9,D7,D8,D10,D6]", "…
#> $ self_reported_ethnicity_ontology_term_id <chr> "HANCESTRO:0005", "HANCESTRO:…
#> $ donor_BMI <chr> "pooled [30.5,22.7,23.5,26.8,…
#> $ donor_times_pregnant <chr> "pooled [3,0,3,2,2]", "pooled…
#> $ family_history_breast_cancer <chr> "pooled [unknown,False,False,…
#> $ organism_ontology_term_id <chr> "NCBITaxon:9606", "NCBITaxon:…
#> $ tyrer_cuzick_lifetime_risk <chr> "pooled [12,14.8,8.8,14.3,20.…
#> $ sample_uuid <chr> "pooled [f008c67a-abb4-4563-8…
#> $ sample_preservation_method <chr> "cryopreservation", "cryopres…
#> $ tissue_ontology_term_id <chr> "UBERON:0035328", "UBERON:003…
#> $ development_stage_ontology_term_id <chr> "HsapDv:0000087", "HsapDv:000…
#> $ suspension_uuid <chr> "38d793cb-d811-4863-aec0-2fa7…
#> $ suspension_type <chr> "cell", "cell", "cell", "cell…
#> $ library_uuid <chr> "385d8d7c-5038-4f0e-b7f3-ec9a…
#> $ assay_ontology_term_id <chr> "EFO:0009922", "EFO:0009922",…
#> $ mapped_reference_annotation <chr> "GENCODE 28", "GENCODE 28", "…
#> $ is_primary_data <lgl> TRUE, TRUE, TRUE, TRUE, TRUE,…
#> $ cell_type_ontology_term_id <chr> "CL:0011026", "CL:0011026", "…
#> $ author_cell_type <chr> "luminal progenitor", "lumina…
#> $ disease_ontology_term_id <chr> "PATO:0000461", "PATO:0000461…
#> $ sex_ontology_term_id <chr> "PATO:0000383", "PATO:0000383…
#> $ nCount_RNA <dbl> 2937, 5495, 5598, 3775, 2146,…
#> $ nFeature_RNA <dbl> 1183, 1827, 2037, 1448, 1027,…
#> $ percent.mito <dbl> 0.02076949, 0.03676069, 0.043…
#> $ seurat_clusters <dbl> 3, 3, 24, 1, 3, 1, 1, 4, 1, 0…
#> $ sample_id <chr> "CMGpool", "CMGpool", "CMGpoo…
#> $ cell_type <chr> "progenitor cell", "progenito…
#> $ assay <chr> "10x 3' v3", "10x 3' v3", "10…
#> $ disease <chr> "normal", "normal", "normal",…
#> $ organism <chr> "Homo sapiens", "Homo sapiens…
#> $ sex <chr> "female", "female", "female",…
#> $ tissue <chr> "upper outer quadrant of brea…
#> $ self_reported_ethnicity <chr> "European", "European", "Euro…
#> $ development_stage <chr> "human adult stage", "human a…Summarize information – how many donors, what developmental stage, what ethnicity?
cell_data |>
count(donor_id, development_stage, self_reported_ethnicity)
#> # A tibble: 7 × 4
#> donor_id development_stage self_reported_ethnicity n
#> <chr> <chr> <chr> <int>
#> 1 D1 35-year-old human stage European 2303
#> 2 D11 43-year-old human stage Chinese 7454
#> 3 D2 60-year-old human stage European 864
#> 4 D3 44-year-old human stage African American 2517
#> 5 D4 42-year-old human stage European 1771
#> 6 D5 21-year-old human stage European 2244
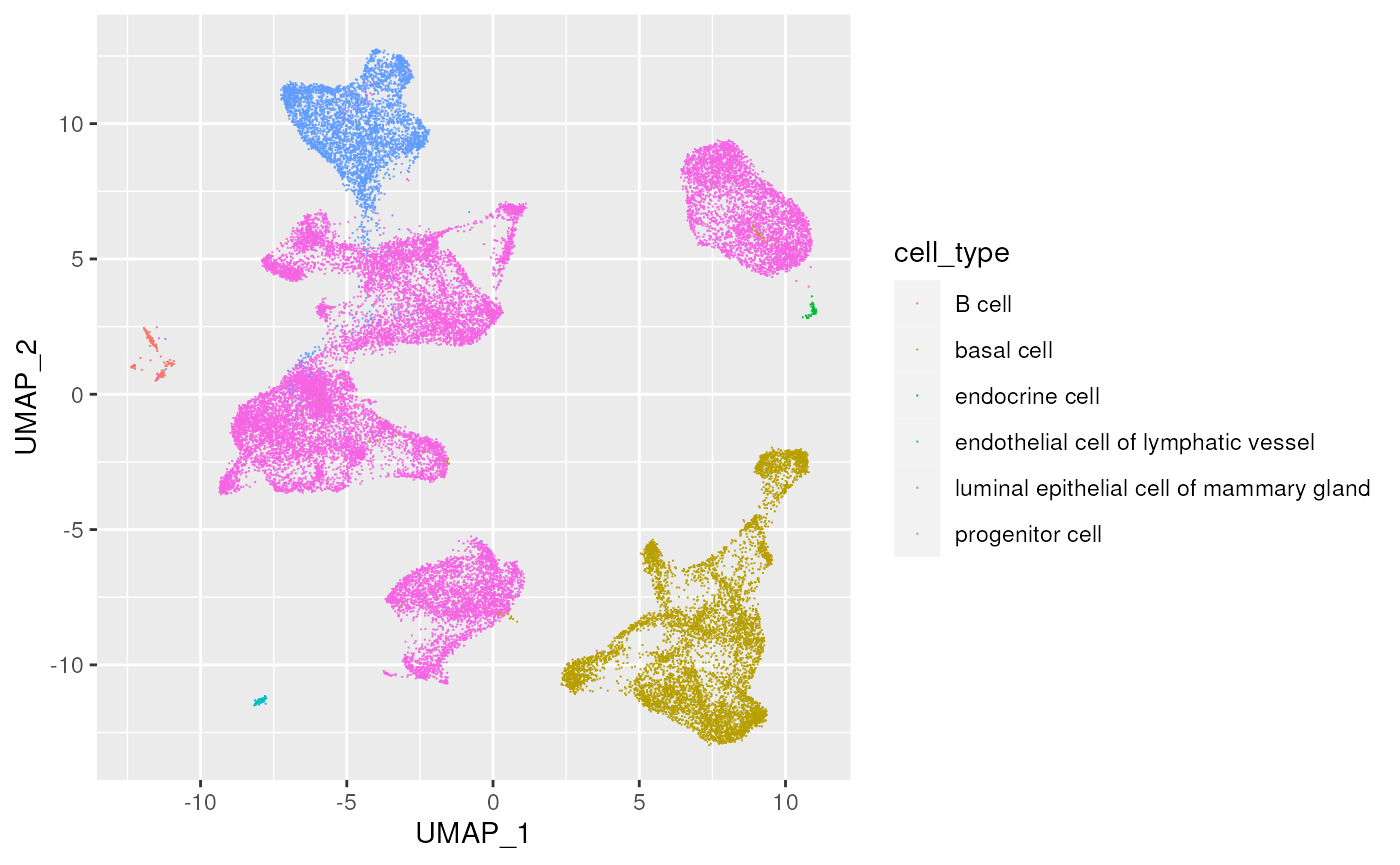
#> 7 pooled [D9,D7,D8,D10,D6] human adult stage European 14543What cell types have been annotated?
cell_data |>
count(cell_type)
#> # A tibble: 6 × 2
#> cell_type n
#> <chr> <int>
#> 1 B cell 215
#> 2 basal cell 7040
#> 3 endocrine cell 64
#> 4 endothelial cell of lymphatic vessel 133
#> 5 luminal epithelial cell of mammary gland 4257
#> 6 progenitor cell 19987Cell types for each ethnicity?
cell_data |>
count(self_reported_ethnicity, cell_type) |>
tidyr::pivot_wider(
names_from = "self_reported_ethnicity",
values_from = "n"
)
#> # A tibble: 6 × 4
#> cell_type `African American` Chinese European
#> <chr> <int> <int> <int>
#> 1 B cell 5 73 137
#> 2 basal cell 583 3367 3090
#> 3 endothelial cell of lymphatic vessel 11 31 91
#> 4 luminal epithelial cell of mammary gland 809 187 3261
#> 5 progenitor cell 1109 3755 15123
#> 6 endocrine cell NA 41 23Reflecting on this – there is no replication across non-European ethnicity, so no statistical insights available. Pooled samples probably require careful treatment in any downstream analysis.
Genes
## use `file.choose()` or similar for your own data sets
row_data_csv <- system.file(package = "ARTNet2023", "scrnaseq-gene-data.csv")
row_data <- readr::read_csv(row_data_csv)
row_data |>
glimpse()
#> Rows: 33,234
#> Columns: 7
#> $ gene_id <chr> "ENSG00000243485", "ENSG00000237613", "ENSG0000018…
#> $ feature_is_filtered <lgl> TRUE, TRUE, TRUE, FALSE, TRUE, TRUE, TRUE, TRUE, T…
#> $ feature_name <chr> "MIR1302-2HG", "FAM138A", "OR4F5", "RP11-34P13.7",…
#> $ feature_reference <chr> "NCBITaxon:9606", "NCBITaxon:9606", "NCBITaxon:960…
#> $ feature_biotype <chr> "gene", "gene", "gene", "gene", "gene", "gene", "g…
#> $ mean_expression <dbl> 3.154972e-05, 0.000000e+00, 0.000000e+00, 2.208481…
#> $ mean_log_expression <dbl> 0.0000218686, 0.0000000000, 0.0000000000, 0.001521…Approximately 1/3rd have been flagged to be filtered. All genes are
from humans (NCBITaxon:9606) and are of biotype ‘gene’.
row_data |>
count(feature_is_filtered, feature_reference, feature_biotype)
#> # A tibble: 2 × 4
#> feature_is_filtered feature_reference feature_biotype n
#> <lgl> <chr> <chr> <int>
#> 1 FALSE NCBITaxon:9606 gene 22743
#> 2 TRUE NCBITaxon:9606 gene 10491A simple plot shows the distribution of log-transformed average expression of each gene
row_data |>
filter(!feature_is_filtered) |>
ggplot() +
aes(x = mean_log_expression) +
geom_histogram()
#> `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
Gene annotations
Introduction to Bioconductor
Web site – https://bioconductor.org
- Available packages https://bioconductor.org/packages
- Package landing pages & vignettes, e.g., https://bioconductor.org/packages/scater
Package installation
- Use CRAN package BiocManager
- Bioconductor, CRAN, and github packages
if (!"BiocManager" %in% rownames(installed.packages()))
install.packages("BiocManager", repos = "https://cran.R-project.org")
BiocManager::install("GenomicFeatures")Support site – https://support.bioconductor.org
- also
- slack – sign up - https://slack.bioconductor.org/
- Bug reports, e.g.,
bug.report(package = "GenomicFeatures") - direct email to maintainers
maintainer("GenomicFeatures")
Source code
-
git clone https://git.bioconductor.org/packages/GenomicFeatures
Other resources
Annotations
From row_data, we know each Ensembl gene id, but what
else can we learn about these genes?
library(AnnotationHub)
ah <- AnnotationHub()
query(ah, c("EnsDb", "Homo sapiens"))
#> AnnotationHub with 24 records
#> # snapshotDate(): 2023-06-23
#> # $dataprovider: Ensembl
#> # $species: Homo sapiens
#> # $rdataclass: EnsDb
#> # additional mcols(): taxonomyid, genome, description,
#> # coordinate_1_based, maintainer, rdatadateadded, preparerclass, tags,
#> # rdatapath, sourceurl, sourcetype
#> # retrieve records with, e.g., 'object[["AH53211"]]'
#>
#> title
#> AH53211 | Ensembl 87 EnsDb for Homo Sapiens
#> AH53715 | Ensembl 88 EnsDb for Homo Sapiens
#> AH56681 | Ensembl 89 EnsDb for Homo Sapiens
#> AH57757 | Ensembl 90 EnsDb for Homo Sapiens
#> AH60773 | Ensembl 91 EnsDb for Homo Sapiens
#> ... ...
#> AH98047 | Ensembl 105 EnsDb for Homo sapiens
#> AH100643 | Ensembl 106 EnsDb for Homo sapiens
#> AH104864 | Ensembl 107 EnsDb for Homo sapiens
#> AH109336 | Ensembl 108 EnsDb for Homo sapiens
#> AH109606 | Ensembl 109 EnsDb for Homo sapiens
ensdb109 <- ah[["AH109606"]]
#> loading from cache
#> require("ensembldb")
ensdb109
#> EnsDb for Ensembl:
#> |Backend: SQLite
#> |Db type: EnsDb
#> |Type of Gene ID: Ensembl Gene ID
#> |Supporting package: ensembldb
#> |Db created by: ensembldb package from Bioconductor
#> |script_version: 0.3.10
#> |Creation time: Thu Feb 16 12:36:05 2023
#> |ensembl_version: 109
#> |ensembl_host: localhost
#> |Organism: Homo sapiens
#> |taxonomy_id: 9606
#> |genome_build: GRCh38
#> |DBSCHEMAVERSION: 2.2
#> |common_name: human
#> |species: homo_sapiens
#> | No. of genes: 70623.
#> | No. of transcripts: 276218.
#> |Protein data available.There are a number of ‘tables’ of data in the EnsDb; check out
browseVignettes("ensembldb") for more information.
names(listTables(ensdb109))
#> [1] "gene" "tx" "tx2exon" "exon"
#> [5] "chromosome" "protein" "uniprot" "protein_domain"
#> [9] "entrezgene" "metadata"E.g., add information about each unfiltered gene from
row_data.
-
get gene annotations from the EnsDB object
gene_annotations <- genes( ensdb109, filter = ~ gene_biotype == "protein_coding", return.type = "DataFrame" ) |> as_tibble() gene_annotations #> # A tibble: 22,907 × 13 #> gene_id gene_name gene_biotype gene_seq_start gene_seq_end seq_name #> <chr> <chr> <chr> <int> <int> <chr> #> 1 ENSG00000186092 OR4F5 protein_coding 65419 71585 1 #> 2 ENSG00000284733 OR4F29 protein_coding 450740 451678 1 #> 3 ENSG00000284662 OR4F16 protein_coding 685716 686654 1 #> 4 ENSG00000187634 SAMD11 protein_coding 923923 944575 1 #> 5 ENSG00000188976 NOC2L protein_coding 944203 959309 1 #> 6 ENSG00000187961 KLHL17 protein_coding 960584 965719 1 #> 7 ENSG00000187583 PLEKHN1 protein_coding 966482 975865 1 #> 8 ENSG00000187642 PERM1 protein_coding 975198 982117 1 #> 9 ENSG00000188290 HES4 protein_coding 998962 1000172 1 #> 10 ENSG00000187608 ISG15 protein_coding 1001138 1014540 1 #> # ℹ 22,897 more rows #> # ℹ 7 more variables: seq_strand <int>, seq_coord_system <chr>, #> # description <chr>, gene_id_version <chr>, canonical_transcript <chr>, #> # symbol <chr>, entrezid <named list> -
left_join()the filteredrow_datatogene_annotations(i.e., keep all rows from the filtered row data, and add columns for matching rows fromgene_annotations)row_data |> dplyr::filter(!feature_is_filtered) |> left_join(gene_annotations) #> Joining with `by = join_by(gene_id)` #> # A tibble: 22,743 × 19 #> gene_id feature_is_filtered feature_name feature_reference feature_biotype #> <chr> <lgl> <chr> <chr> <chr> #> 1 ENSG00000… FALSE RP11-34P13.7 NCBITaxon:9606 gene #> 2 ENSG00000… FALSE LINC01409 NCBITaxon:9606 gene #> 3 ENSG00000… FALSE FAM87B NCBITaxon:9606 gene #> 4 ENSG00000… FALSE LINC00115 NCBITaxon:9606 gene #> 5 ENSG00000… FALSE FAM41C NCBITaxon:9606 gene #> 6 ENSG00000… FALSE RP11-54O7.1 NCBITaxon:9606 gene #> 7 ENSG00000… FALSE LINC02593 NCBITaxon:9606 gene #> 8 ENSG00000… FALSE SAMD11 NCBITaxon:9606 gene #> 9 ENSG00000… FALSE NOC2L NCBITaxon:9606 gene #> 10 ENSG00000… FALSE KLHL17 NCBITaxon:9606 gene #> # ℹ 22,733 more rows #> # ℹ 14 more variables: mean_expression <dbl>, mean_log_expression <dbl>, #> # gene_name <chr>, gene_biotype <chr>, gene_seq_start <int>, #> # gene_seq_end <int>, seq_name <chr>, seq_strand <int>, #> # seq_coord_system <chr>, description <chr>, gene_id_version <chr>, #> # canonical_transcript <chr>, symbol <chr>, entrezid <named list>
Many other annotation resources available, to help place information about genes into biological context.
SingleCellExperiment
Row (gene) data, column (cell) data, and a matrix of counts describe
a single cell experiment. These can be assembled, along with other
information about, e.g., reduced dimension representations, into a
‘SingleCellExperiment’ Bioconductor object (see
?SingleCellExperiment).
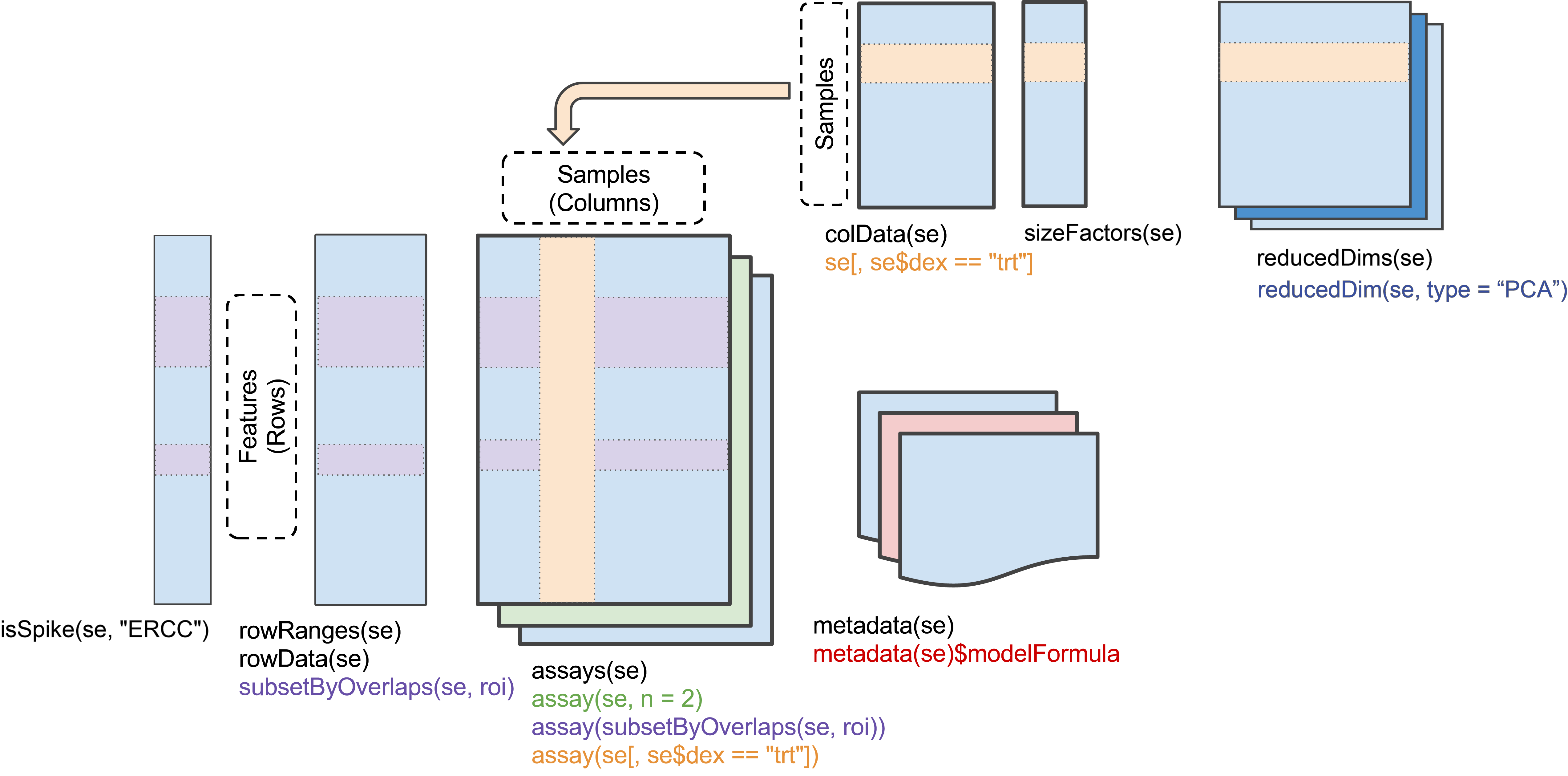
Here we illustrate this construction with some artificial data:
library(SingleCellExperiment)
n_genes <- 200
n_cells <- 100
demo_count <- matrix(rpois(20000, 5), ncol=n_cells) # counts
demo_log_count <- log2(demo_count + 1) # log counts
demo_row_data <- data.frame(
gene_id = paste0("gene_", seq_len(n_genes))
)
demo_column_data <- data.frame(
cell_id = paste0("cell_", seq_len(n_cells))
)
demo_pca <- matrix(runif(n_cells * 5), n_cells)
demo_tsne <- matrix(rnorm(n_cells * 2), n_cells)
demo_sce <- SingleCellExperiment(
assays=list(counts=demo_count, logcounts=demo_log_count),
colData = demo_column_data,
rowData = demo_row_data,
reducedDims=SimpleList(PCA=demo_pca, tSNE=demo_tsne)
)
demo_sce
#> class: SingleCellExperiment
#> dim: 200 100
#> metadata(0):
#> assays(2): counts logcounts
#> rownames: NULL
#> rowData names(1): gene_id
#> colnames: NULL
#> colData names(1): cell_id
#> reducedDimNames(2): PCA tSNE
#> mainExpName: NULL
#> altExpNames(0):Elements of the object can be obtained using ’accessors`, e.g.,
Summary
Session information
This document was produced with the following R software:
sessionInfo()
#> R version 4.3.0 (2023-04-21)
#> Platform: x86_64-pc-linux-gnu (64-bit)
#> Running under: Ubuntu 22.04.2 LTS
#>
#> Matrix products: default
#> BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
#> LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.20.so; LAPACK version 3.10.0
#>
#> locale:
#> [1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
#> [3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
#> [5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
#> [7] LC_PAPER=en_US.UTF-8 LC_NAME=C
#> [9] LC_ADDRESS=C LC_TELEPHONE=C
#> [11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
#>
#> time zone: Etc/UTC
#> tzcode source: system (glibc)
#>
#> attached base packages:
#> [1] stats4 stats graphics grDevices utils datasets methods
#> [8] base
#>
#> other attached packages:
#> [1] SingleCellExperiment_1.23.0 SummarizedExperiment_1.31.1
#> [3] MatrixGenerics_1.13.0 matrixStats_1.0.0
#> [5] ensembldb_2.25.0 AnnotationFilter_1.25.0
#> [7] GenomicFeatures_1.53.1 AnnotationDbi_1.63.1
#> [9] Biobase_2.61.0 GenomicRanges_1.53.1
#> [11] GenomeInfoDb_1.37.2 IRanges_2.35.2
#> [13] S4Vectors_0.39.1 AnnotationHub_3.9.1
#> [15] BiocFileCache_2.9.0 dbplyr_2.3.2
#> [17] BiocGenerics_0.47.0 plotly_4.10.2
#> [19] ggplot2_3.4.2 dplyr_1.1.2
#>
#> loaded via a namespace (and not attached):
#> [1] jsonlite_1.8.5 magrittr_2.0.3
#> [3] farver_2.1.1 rmarkdown_2.22
#> [5] fs_1.6.2 BiocIO_1.11.0
#> [7] zlibbioc_1.47.0 ragg_1.2.5
#> [9] vctrs_0.6.3 memoise_2.0.1
#> [11] Rsamtools_2.17.0 RCurl_1.98-1.12
#> [13] htmltools_0.5.5 S4Arrays_1.1.4
#> [15] progress_1.2.2 curl_5.0.1
#> [17] SparseArray_1.1.10 sass_0.4.6
#> [19] bslib_0.5.0 htmlwidgets_1.6.2
#> [21] desc_1.4.2 cachem_1.0.8
#> [23] GenomicAlignments_1.37.0 mime_0.12
#> [25] lifecycle_1.0.3 pkgconfig_2.0.3
#> [27] Matrix_1.5-4.1 R6_2.5.1
#> [29] fastmap_1.1.1 GenomeInfoDbData_1.2.10
#> [31] shiny_1.7.4 digest_0.6.32
#> [33] colorspace_2.1-0 rprojroot_2.0.3
#> [35] textshaping_0.3.6 crosstalk_1.2.0
#> [37] RSQLite_2.3.1 filelock_1.0.2
#> [39] labeling_0.4.2 fansi_1.0.4
#> [41] httr_1.4.6 compiler_4.3.0
#> [43] bit64_4.0.5 withr_2.5.0
#> [45] BiocParallel_1.35.2 DBI_1.1.3
#> [47] highr_0.10 biomaRt_2.57.1
#> [49] rappdirs_0.3.3 DelayedArray_0.27.5
#> [51] rjson_0.2.21 tools_4.3.0
#> [53] interactiveDisplayBase_1.39.0 httpuv_1.6.11
#> [55] glue_1.6.2 restfulr_0.0.15
#> [57] promises_1.2.0.1 grid_4.3.0
#> [59] generics_0.1.3 gtable_0.3.3
#> [61] tzdb_0.4.0 tidyr_1.3.0
#> [63] data.table_1.14.8 hms_1.1.3
#> [65] xml2_1.3.4 utf8_1.2.3
#> [67] XVector_0.41.1 BiocVersion_3.18.0
#> [69] pillar_1.9.0 stringr_1.5.0
#> [71] vroom_1.6.3 later_1.3.1
#> [73] lattice_0.21-8 rtracklayer_1.61.0
#> [75] bit_4.0.5 tidyselect_1.2.0
#> [77] Biostrings_2.69.1 knitr_1.43
#> [79] ProtGenerics_1.33.1 xfun_0.39
#> [81] stringi_1.7.12 lazyeval_0.2.2
#> [83] yaml_2.3.7 evaluate_0.21
#> [85] codetools_0.2-19 tibble_3.2.1
#> [87] BiocManager_1.30.21 cli_3.6.1
#> [89] xtable_1.8-4 systemfonts_1.0.4
#> [91] munsell_0.5.0 jquerylib_0.1.4
#> [93] Rcpp_1.0.10 png_0.1-8
#> [95] XML_3.99-0.14 parallel_4.3.0
#> [97] ellipsis_0.3.2 pkgdown_2.0.7
#> [99] readr_2.1.4 blob_1.2.4
#> [101] prettyunits_1.1.1 bitops_1.0-7
#> [103] viridisLite_0.4.2 scales_1.2.1
#> [105] purrr_1.0.1 crayon_1.5.2
#> [107] rlang_1.1.1 KEGGREST_1.41.0