Example data: BRFSS
We use data from the US Center for Disease Control’s Behavioral Risk Factor Surveillance System (BRFSS) annual survey. Check out the web page for a little more information. We are using a small subset of this data, including a random sample of 20000 observations from each of 1990 and 2010.
Data input
From Excel to CSV
Getting a dataset into R is a necessary and sometimes tedious process. Often data starts as an Excel spreadsheet…
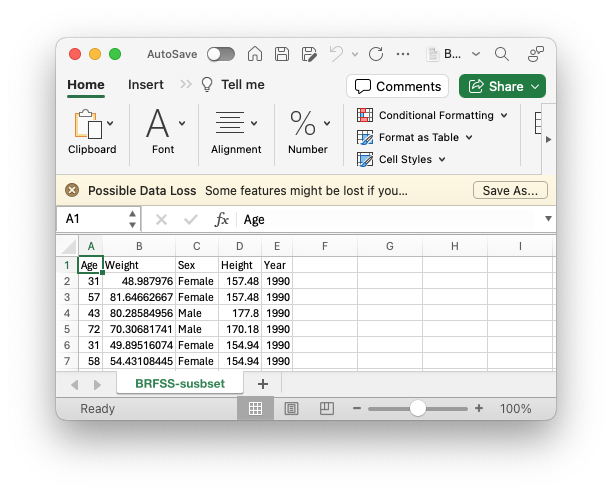
Note that this is a straight-forward sheet – the first row provides a column name. Each subsequent row corresponds to a single observation. There are no mouse-over ‘comments’ or secondary tables or… This simple structure is a good starting point for moving data to R.
Our first step is to export the data from Excel to a ‘CSV’ file. We do this because (a) CSV files are easily read into R and (b) many other data sources can be transformed to CSV format. Chose the ‘File’ menu and then ‘Save’. On macOS I then chose to save the data as CSV with a dialog box that looked like…
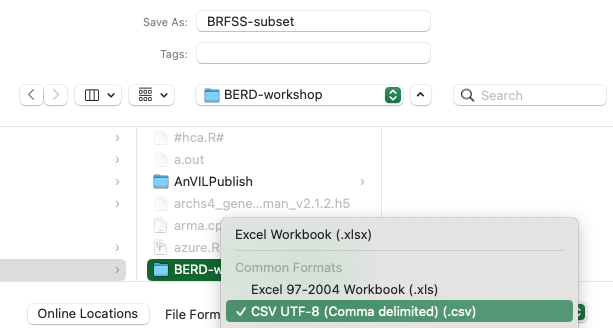
This procedure would create a file ‘BRFSS-subset.csv’ on our computer.
We can’t start with an Excel file, so instead download the data we
will work with using the following command. You might need to make sure
that the folder ‘BRFSS-workshop’ exists at the file system location
returned by getwd().
getwd()
url <- "https://raw.githubusercontent.com/mtmorgan/BERD/main/inst/extdata/BRFSS-subset.csv"
destination <- file.path("BRFSS-workshop", basename(url))
if (!file.exists(destination)) {
dir.create("data")
download.file(url, destination)
}CSV to R
Read the data into R as a tibble using the readr package.
brfss <- readr::read_csv(destination)
#> Rows: 20000 Columns: 5
#> ── Column specification ────────────────────────────────────────────────────────
#> Delimiter: ","
#> chr (1): Sex
#> dbl (4): Age, Weight, Height, Year
#>
#> ℹ Use `spec()` to retrieve the full column specification for this data.
#> ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
brfss
#> # A tibble: 20,000 × 5
#> Age Weight Sex Height Year
#> <dbl> <dbl> <chr> <dbl> <dbl>
#> 1 31 49.0 Female 157. 1990
#> 2 57 81.6 Female 157. 1990
#> 3 43 80.3 Male 178. 1990
#> 4 72 70.3 Male 170. 1990
#> 5 31 49.9 Female 155. 1990
#> 6 58 54.4 Female 155. 1990
#> 7 45 69.9 Male 173. 1990
#> 8 37 68.0 Male 180. 1990
#> 9 33 65.8 Female 170. 1990
#> 10 75 70.8 Female 152. 1990
#> # ℹ 19,990 more rowsExplore the data using dplyr
Attach dplyr to our current R session.
Useful functions for data exploration
-
count()– count occurence of values in one or more columns -
summarize()– more flexible summary of columns -
group_by()– summarize or perform other operations by group
Functions for data manipulation
-
mutate()– update or add columns -
filter()– remove rows based on column values -
select()– remove, rearrange, or re-name columns
count()
Count of observed values for one…
brfss |>
count(Year)
#> # A tibble: 2 × 2
#> Year n
#> <dbl> <int>
#> 1 1990 10000
#> 2 2010 10000
brfss |>
count(Sex)
#> # A tibble: 2 × 2
#> Sex n
#> <chr> <int>
#> 1 Female 12039
#> 2 Male 7961…or several columns
brfss |>
count(Year, Sex)
#> # A tibble: 4 × 3
#> Year Sex n
#> <dbl> <chr> <int>
#> 1 1990 Female 5718
#> 2 1990 Male 4282
#> 3 2010 Female 6321
#> 4 2010 Male 3679
summarize()
More flexible summary, e.g., number of observations n()
or mean values of columns, removing NA,
mean(, na.rm = TRUE).
group_by()
Often data sets contain groups that can be summarized separately, e.g., calculating mean values by Year and Sex.
brfss |>
group_by(Year, Sex) |>
summarize(
n = n(),
ave_age = mean(Age, na.rm = TRUE),
ave_wt = mean(Weight, na.rm = TRUE),
ave_ht = mean(Height, na.rm = TRUE)
)
#> `summarise()` has grouped output by 'Year'. You can override using the
#> `.groups` argument.
#> # A tibble: 4 × 6
#> # Groups: Year [2]
#> Year Sex n ave_age ave_wt ave_ht
#> <dbl> <chr> <int> <dbl> <dbl> <dbl>
#> 1 1990 Female 5718 46.2 64.8 163.
#> 2 1990 Male 4282 43.9 81.2 178.
#> 3 2010 Female 6321 57.1 73.0 163.
#> 4 2010 Male 3679 56.2 88.8 178.
mutate()
Year and Sex should really be factors.
Also, add a column for log-10 transformed weight.
brfss_clean <-
brfss |>
mutate(
Year = factor(Year, levels = c("1990", "2010")),
Sex = factor(Sex, levels = c("Female", "Male")),
Log10Weight = log10(Weight)
)
brfss_clean
#> # A tibble: 20,000 × 6
#> Age Weight Sex Height Year Log10Weight
#> <dbl> <dbl> <fct> <dbl> <fct> <dbl>
#> 1 31 49.0 Female 157. 1990 1.69
#> 2 57 81.6 Female 157. 1990 1.91
#> 3 43 80.3 Male 178. 1990 1.90
#> 4 72 70.3 Male 170. 1990 1.85
#> 5 31 49.9 Female 155. 1990 1.70
#> 6 58 54.4 Female 155. 1990 1.74
#> 7 45 69.9 Male 173. 1990 1.84
#> 8 37 68.0 Male 180. 1990 1.83
#> 9 33 65.8 Female 170. 1990 1.82
#> 10 75 70.8 Female 152. 1990 1.85
#> # ℹ 19,990 more rows
filter()
Create a subset of the data that includes only Male samples, …
brfss_male <-
brfss_clean |>
filter(Sex == "Male")
brfss_male
#> # A tibble: 7,961 × 6
#> Age Weight Sex Height Year Log10Weight
#> <dbl> <dbl> <fct> <dbl> <fct> <dbl>
#> 1 43 80.3 Male 178. 1990 1.90
#> 2 72 70.3 Male 170. 1990 1.85
#> 3 45 69.9 Male 173. 1990 1.84
#> 4 37 68.0 Male 180. 1990 1.83
#> 5 56 88.5 Male 180. 1990 1.95
#> 6 74 81.6 Male 183. 1990 1.91
#> 7 19 93.0 Male 183. 1990 1.97
#> 8 35 97.5 Male 193. 1990 1.99
#> 9 60 78.0 Male 170. 1990 1.89
#> 10 29 77.1 Male 175. 1990 1.89
#> # ℹ 7,951 more rows…or only Female samples from 2010.
brfss_female_2010 <-
brfss_clean |>
filter(Sex == "Female", Year == "2010")
brfss_female_2010
#> # A tibble: 6,321 × 6
#> Age Weight Sex Height Year Log10Weight
#> <dbl> <dbl> <fct> <dbl> <fct> <dbl>
#> 1 46 45.4 Female 165 2010 1.66
#> 2 49 145. Female 183 2010 2.16
#> 3 53 54.4 Female 155 2010 1.74
#> 4 63 77.1 Female 170 2010 1.89
#> 5 53 82.1 Female 168 2010 1.91
#> 6 64 90.7 Female 163 2010 1.96
#> 7 49 77.1 Female 173 2010 1.89
#> 8 77 72.6 Female 168 2010 1.86
#> 9 70 79.4 Female 178 2010 1.90
#> 10 83 93.0 Female 157 2010 1.97
#> # ℹ 6,311 more rows
select()
Use select() to choose particular columns, or to
re-order columns
brfss_male |>
select(Year, Age, Weight, Log10Weight, Height)
#> # A tibble: 7,961 × 5
#> Year Age Weight Log10Weight Height
#> <fct> <dbl> <dbl> <dbl> <dbl>
#> 1 1990 43 80.3 1.90 178.
#> 2 1990 72 70.3 1.85 170.
#> 3 1990 45 69.9 1.84 173.
#> 4 1990 37 68.0 1.83 180.
#> 5 1990 56 88.5 1.95 180.
#> 6 1990 74 81.6 1.91 183.
#> 7 1990 19 93.0 1.97 183.
#> 8 1990 35 97.5 1.99 193.
#> 9 1990 60 78.0 1.89 170.
#> 10 1990 29 77.1 1.89 175.
#> # ℹ 7,951 more rowsInitial visualization
What is the relationship between Height and Weight in Female samples from 2010?
plot(Weight ~ Height, brfss_female_2010)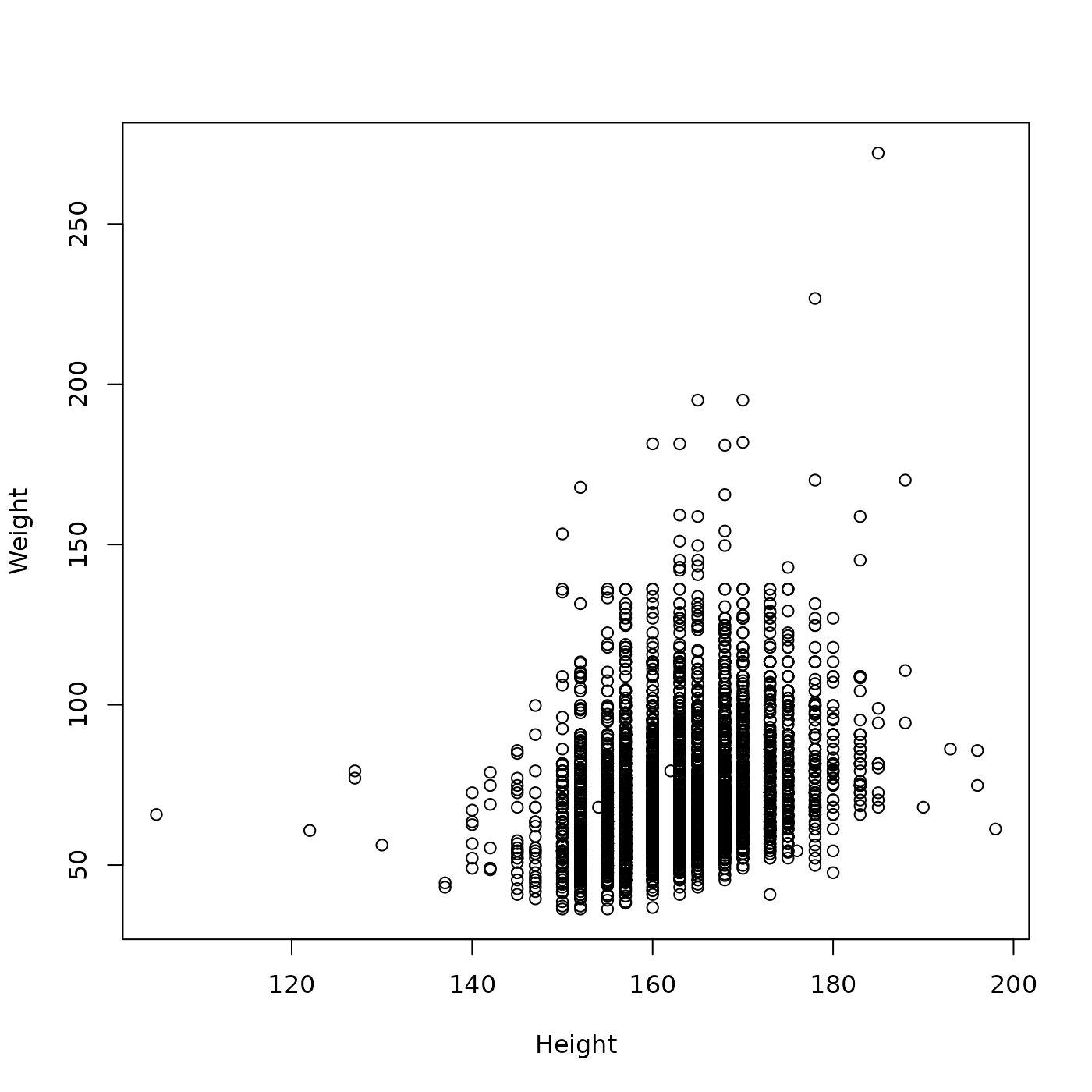
With Weight on a log scale?
plot(Weight ~ Height, brfss_female_2010, log = "y")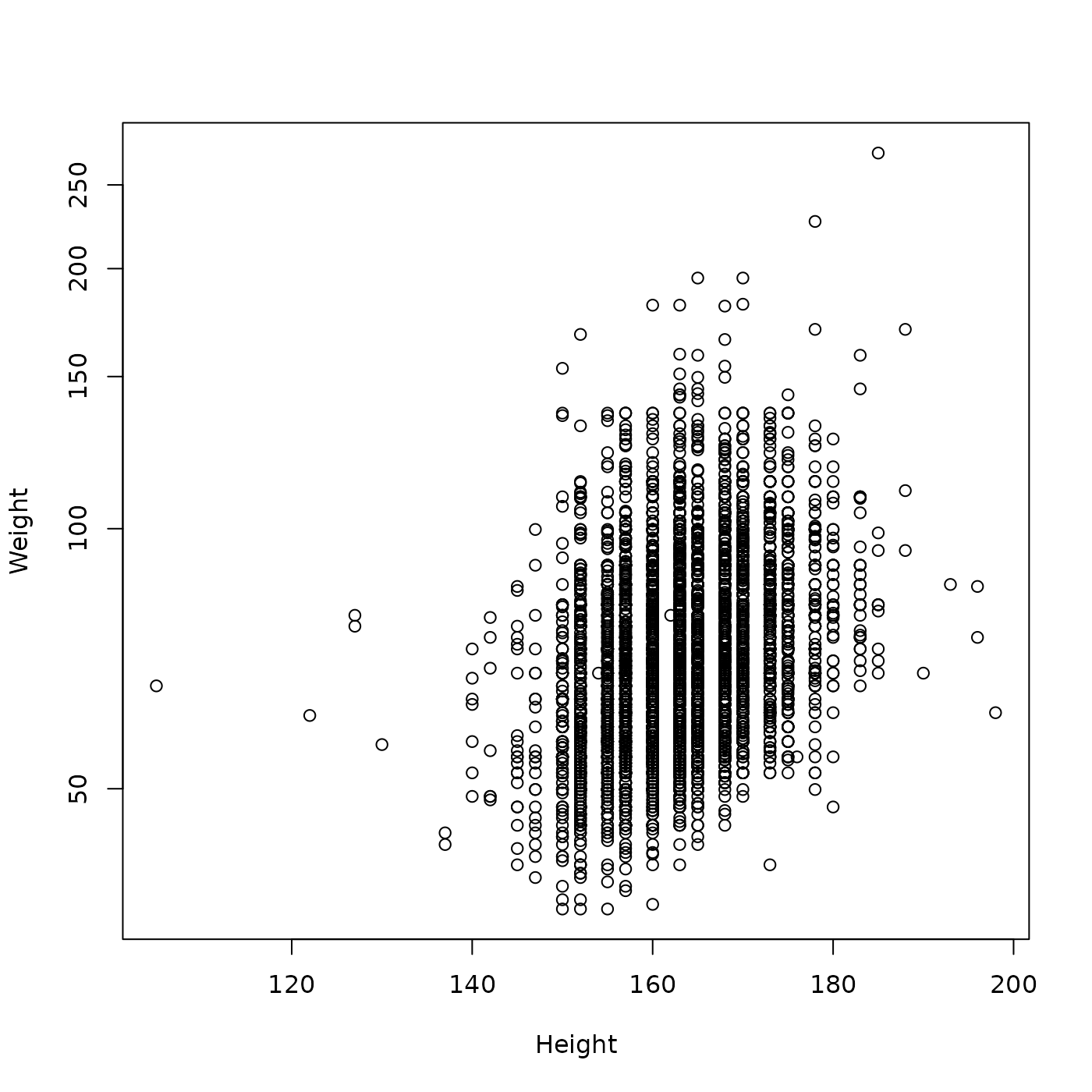
## another way, but maybe the Y-axis scale is harder to interpret?
## plot(Log10Weight ~ Height, brfss_female_2010)Does it look like Males differ in weight between 1990 and 2010, plotted on a log scale?
brfss_male |>
group_by(Year) |>
summarize(
n = n(),
ave_wt = mean(Weight, na.rm = TRUE),
ave_log10_wt = mean(Log10Weight, na.rm = TRUE)
)
#> # A tibble: 2 × 4
#> Year n ave_wt ave_log10_wt
#> <fct> <int> <dbl> <dbl>
#> 1 1990 4282 81.2 1.90
#> 2 2010 3679 88.8 1.94
plot(Weight ~ Year, brfss_male, log = "y")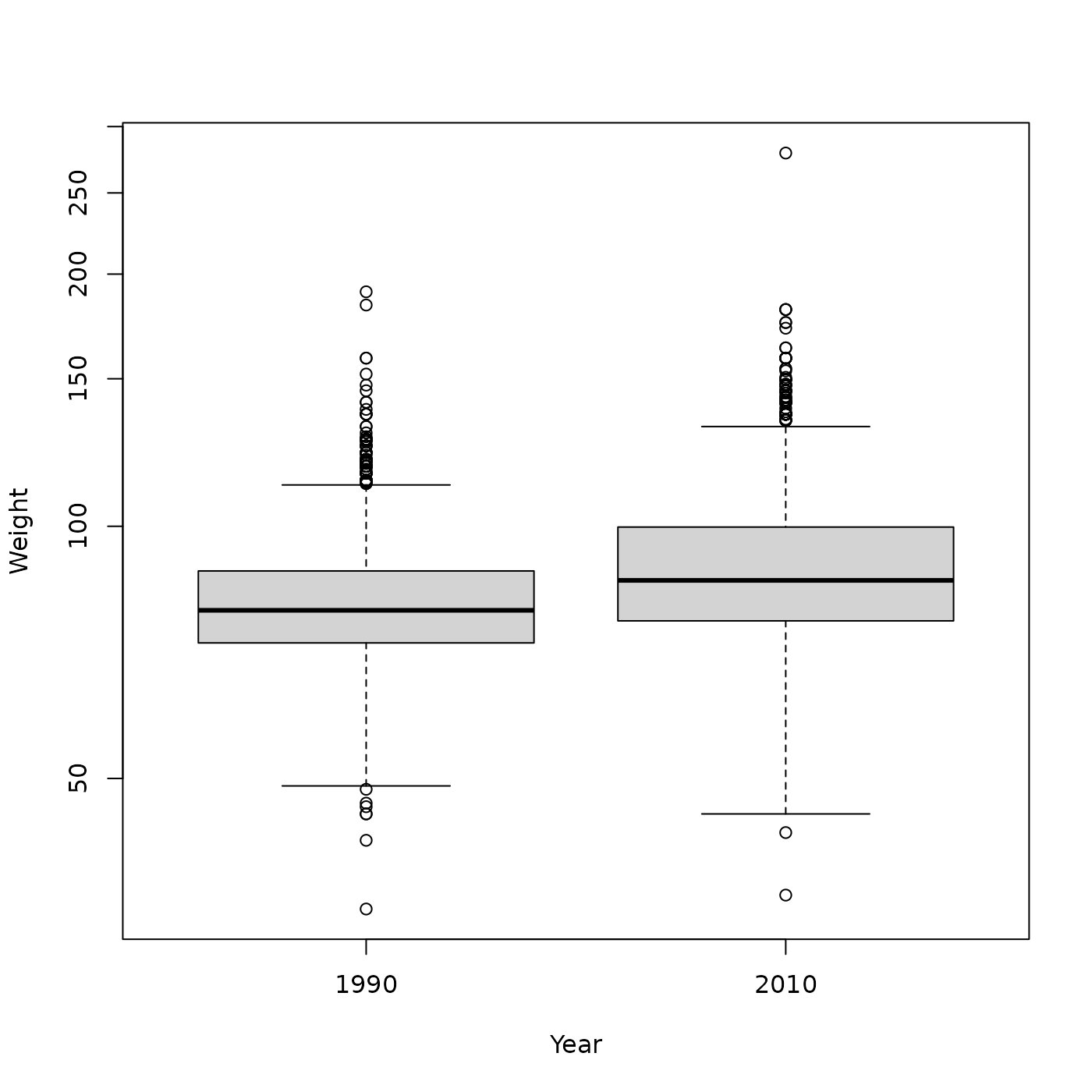
Session information
sessionInfo()
#> R version 4.3.3 (2024-02-29)
#> Platform: x86_64-pc-linux-gnu (64-bit)
#> Running under: Ubuntu 22.04.4 LTS
#>
#> Matrix products: default
#> BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
#> LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.20.so; LAPACK version 3.10.0
#>
#> locale:
#> [1] LC_CTYPE=C.UTF-8 LC_NUMERIC=C LC_TIME=C.UTF-8
#> [4] LC_COLLATE=C.UTF-8 LC_MONETARY=C.UTF-8 LC_MESSAGES=C.UTF-8
#> [7] LC_PAPER=C.UTF-8 LC_NAME=C LC_ADDRESS=C
#> [10] LC_TELEPHONE=C LC_MEASUREMENT=C.UTF-8 LC_IDENTIFICATION=C
#>
#> time zone: UTC
#> tzcode source: system (glibc)
#>
#> attached base packages:
#> [1] stats graphics grDevices utils datasets methods base
#>
#> other attached packages:
#> [1] dplyr_1.1.4
#>
#> loaded via a namespace (and not attached):
#> [1] bit_4.0.5 jsonlite_1.8.8 highr_0.10 compiler_4.3.3
#> [5] crayon_1.5.2 tidyselect_1.2.1 parallel_4.3.3 jquerylib_0.1.4
#> [9] systemfonts_1.0.6 textshaping_0.3.7 yaml_2.3.8 fastmap_1.1.1
#> [13] readr_2.1.5 R6_2.5.1 generics_0.1.3 knitr_1.45
#> [17] tibble_3.2.1 desc_1.4.3 bslib_0.6.2 pillar_1.9.0
#> [21] tzdb_0.4.0 rlang_1.1.3 utf8_1.2.4 cachem_1.0.8
#> [25] xfun_0.43 fs_1.6.3 sass_0.4.9 bit64_4.0.5
#> [29] memoise_2.0.1 cli_3.6.2 withr_3.0.0 pkgdown_2.0.7
#> [33] magrittr_2.0.3 digest_0.6.35 vroom_1.6.5 hms_1.1.3
#> [37] lifecycle_1.0.4 vctrs_0.6.5 evaluate_0.23 glue_1.7.0
#> [41] ragg_1.3.0 fansi_1.0.6 rmarkdown_2.26 purrr_1.0.2
#> [45] tools_4.3.3 pkgconfig_2.0.3 htmltools_0.5.8