A. Introduction to R
a-intro-to-r.RmdEssential R
A simple calculator
1 + 1
## [1] 2‘Vectors’ as building blocks
c(1, 2, 3)
## [1] 1 2 3
c("January", "February", "March")
## [1] "January" "February" "March"
c(TRUE, FALSE)
## [1] TRUE FALSEVariables, missing values and ‘factors’
age <- c(27, NA, 32, 29)
gender <- factor(
c("Female", "Male", "Non-binary", NA),
levels = c("Female", "Male", "Non-binary")
)Data structures to coordinate related vectors – the
data.frame
df <- data.frame(
age = c(27, NA, 32, 29),
gender = gender
)
df
## age gender
## 1 27 Female
## 2 NA Male
## 3 32 Non-binary
## 4 29 <NA>Key opererations on data.frame
-
df[1:3, c("gender", "age")]– subset on rows and columns -
df[["age"]],df$age– select columns
Functions
rnorm(5) # 5 random normal deviates
## [1] 2.79022372 -0.38764450 -0.23800823 2.07697779 -0.04142712
x <- rnorm(100) # 100 random normal deviates
hist(x) # histogram, approximately normal

‘Vectorized’ operations, e.g., element-wise addition without an explicit ‘for’ loop
y <- x + rnorm(100)
plot(y ~ x)
fit <- lm(y ~ x)
fit # an R 'object' containing information about the
##
## Call:
## lm(formula = y ~ x)
##
## Coefficients:
## (Intercept) x
## -0.1138 0.9889
# regression of y on x
abline(fit) # plot points and fitted regression line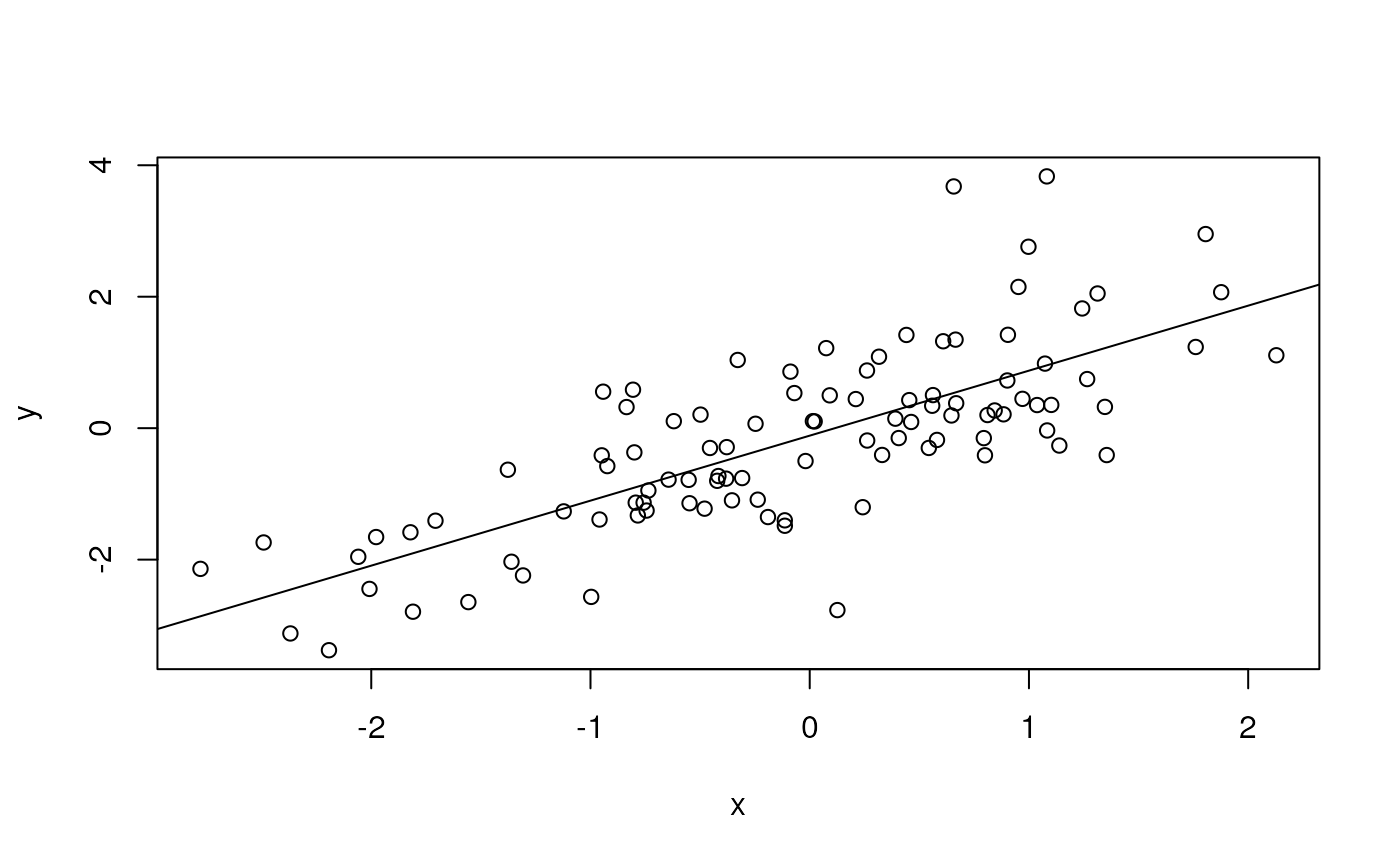
anova(fit) # statistical summary of linear regression
## Analysis of Variance Table
##
## Response: y
## Df Sum Sq Mean Sq F value Pr(>F)
## x 1 107.594 107.594 126.5 < 2.2e-16 ***
## Residuals 98 83.351 0.851
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Write your own functions
hello <- function(who) {
paste("hello", who, "with", nchar(who), "letters in your name")
}
hello("Martin")
## [1] "hello Martin with 6 letters in your name"Iterate, usually with lapply() although
for() is available
Packages
Extend functionality of base R. Can be part of the ‘base’ distribution…
## iterate over the numbers 1 through 8, 'sleeping' for 1 second
## each. Takes about 8 seconds...
system.time({
lapply(1:8, function(i) Sys.sleep(1))
})
## user system elapsed
## 0.003 0.001 8.011
## sleep in parallel -- takes only 2 seconds
library(parallel)
cl <- makeCluster(4) # cluster of 4 workers
system.time({
parLapply(cl, 1:8, function(i) Sys.sleep(1))
})
## user system elapsed
## 0.003 0.000 2.089Tidyverse
The dplyr package introduces the ‘tidyverse’
A ‘tibble’ is like a ‘data.frame’, but more user-friendly
tbl <- tibble(
x = rnorm(100),
y = x + rnorm(100)
)
## e.g., only displays the first 10 rows
tbl
## # A tibble: 100 × 2
## x y
## <dbl> <dbl>
## 1 0.579 1.74
## 2 0.302 -0.214
## 3 -1.51 -2.21
## 4 0.732 0.980
## 5 -1.11 -2.42
## 6 1.36 2.63
## 7 -0.443 -0.989
## 8 -0.391 -0.436
## 9 -1.17 -2.56
## 10 -1.48 -1.72
## # … with 90 more rowsThe tidyverse makes use of ‘pipes’ |> (the older
syntax is %>%). A pipe takes the left-hand side and pass
through to the right-hand side. Key dplyr ‘verbs’ can be
piped together
-
filter()rows -
select()columns -
mutate()to change values -
group_by()operate on groups of rows -
left_join()(and friends) for joining tibbles based on shared columns
tbl |>
## e.g., just rows with non-negative values of x and y
filter(x > 0, y > 0) |>
## add a column
mutate(distance_from_origin = sqrt(x^2 + y^2))
## # A tibble: 28 × 3
## x y distance_from_origin
## <dbl> <dbl> <dbl>
## 1 0.579 1.74 1.83
## 2 0.732 0.980 1.22
## 3 1.36 2.63 2.96
## 4 0.997 1.38 1.71
## 5 1.05 0.974 1.43
## 6 0.195 0.416 0.459
## 7 0.208 0.305 0.369
## 8 0.298 0.819 0.871
## 9 1.24 2.07 2.42
## 10 0.499 0.884 1.02
## # … with 18 more rowsVisualization
Another example of a contributed package is ggplot2 for visualization
library(ggplot2)
ggplot(tbl) +
aes(x, y) + # use 'x' and 'y' columns for plotting...
geom_point() + # ...plot points...
geom_smooth(method = "lm") # ...linear regresion
## `geom_smooth()` using formula = 'y ~ x'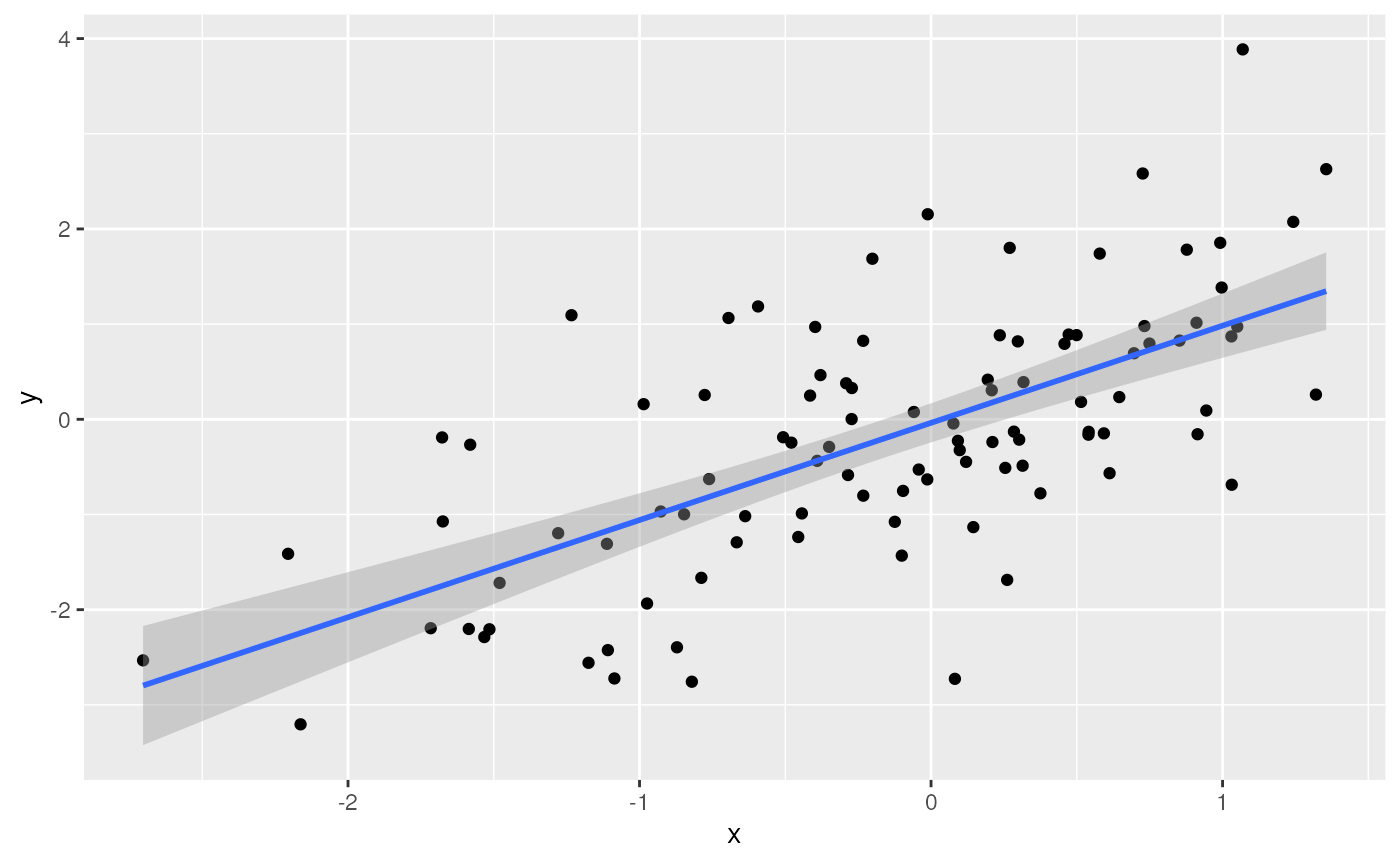
Where do Packages Come From?
CRAN: Comprehensive R Archive Network. More than 18,000 packages. Some help from CRAN Task Views in identifying relevant packages.
Bioconductor: More than 2100 packages relevant to high-throughput genomic analysis. Vignettes are an important part of Bioconductor packages.
Install packages once per R installation, using
BiocManager::install(<package-name>) (CRAN or
Bioconductor)
What about GitHub? Packages haven’t been checked by a formal system, so may have incomplete code, documentation, dependencies on other packages, etc. Authors may not yet be committed to long-term maintenance of their package.