Compiled: 2023-12-12
Note that this is way outside my area of expertise, so I have undoubtedly made terrible blunders.
Introduction
The ‘grantpubcite’ package can be used to query the NIH Reporter database for funded grants, and the publications associated with those grants. The citation history of publications can be discovered using iCite.
In this case study we look at the Immuno-Oncology Translational Network (IOTN), a consortium supported by the NIH Cancer Moonshot to accelerate translation of basic discoveries to clinical applications to improve immunotherapy outcomes.
Getting started
See the Introduction to ‘grantpubcite’ article for installation, basic use, and a brief introduction to ‘tidyverse’ operations.
Load the library and other packages to be used in this case study.
This document is written in Rmarkdown; code chunks used to generate each table or figure can be shown by toggling the ‘Details’ widget.
Projects
Funding Opportunity Announcements for IOTN are as follows:
foas <- case_study_foa_iotn()Discover projects funded through these FOAs by querying NIH Reporter, and use the information returned by a the query to summarize the projects.
## not all projects funded by these FOAs are part of the IOTN
excluded_core_project_num <- c(
"U01CA247548", "U01CA247576", "UH3CA244687", "UH3CA244697"
)
project_summary <-
program_projects(foas) |>
filter(!(core_project_num %in% excluded_core_project_num))There are 31 projects. The projects include large U54, resource sharing and management centers, as well as U01 and UG3 awards.
plot <-
project_summary |>
left_join(foas, by = "opportunity_number") |>
ggplot(aes(tag, award_amount)) +
scale_y_continuous(labels = scales::comma) +
geom_boxplot(outlier.color = NA) +
geom_jitter() +
xlab("Funding type") + ylab("Amount ($)")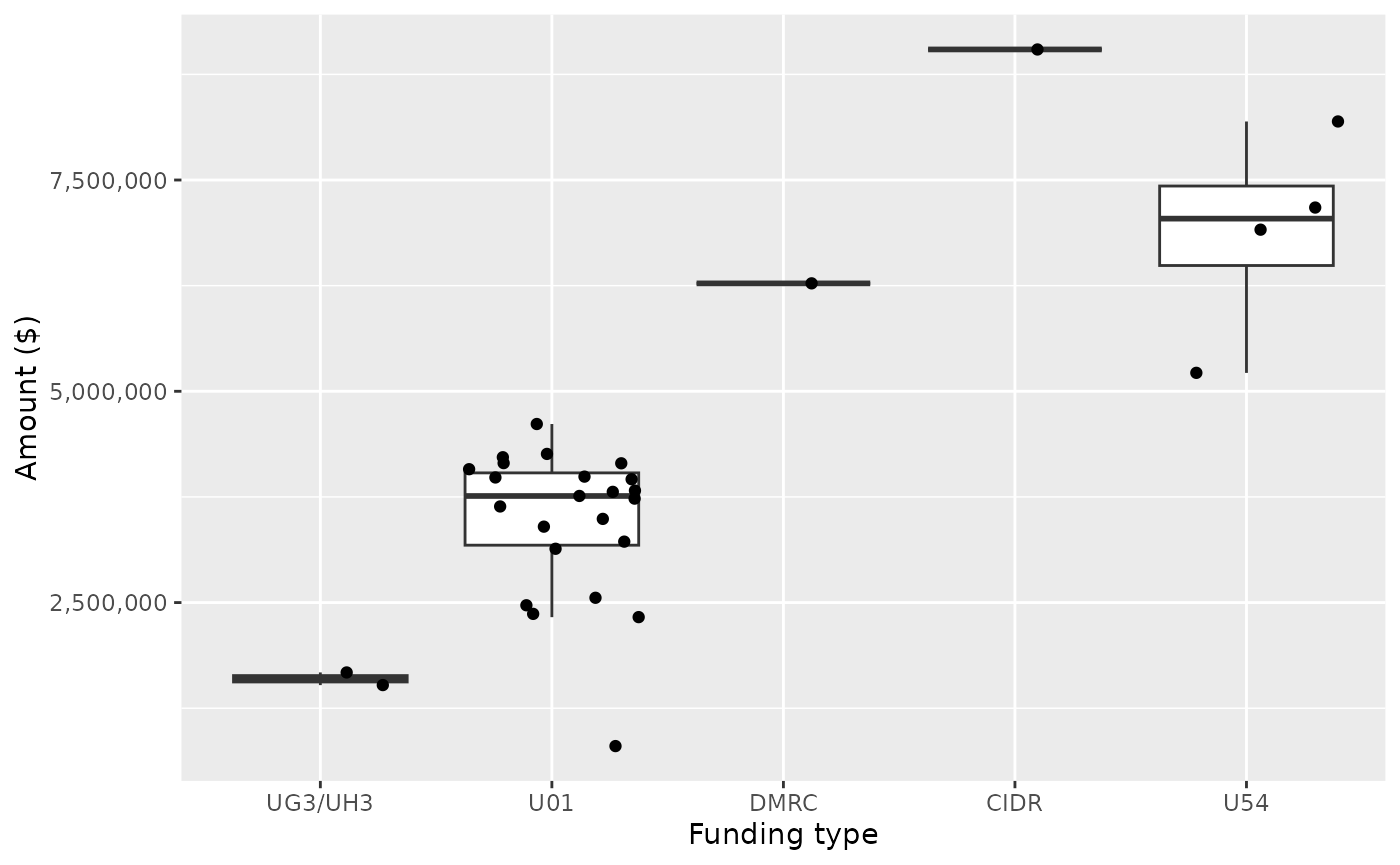
Publications and citations
Publications associated with projects are discovered using NIH Reporter. Citations are from iCite.
program_publications <-
program_publications(foas) |>
filter(!(core_project_num %in% excluded_core_project_num))
publications <-
program_publications |>
select(-c("opportunity_number", "core_project_num")) |>
distinct() |>
arrange(desc(citation_count))
publications_per_project <-
program_publications |>
count(core_project_num, name = "n_pub") |>
left_join(project_summary, by = "core_project_num") |>
select(-c("opportunity_number", "project_title", "fiscal_year")) |>
arrange(desc(n_pub))
collaboration_summary <-
program_publications |>
count(pmid, name = "n_collaborators") |>
count(n_collaborators, name = "n_publications")There are 500 publications. Project U01DK124165 had a very important publication with PMID 32839624.
About 13% of publications involved collaboration between projects; one publication represented an extensive collaboration.
The network below show collaborations between projects, excluding the extensive colloration. Hover over nodes to see project number and title. The width of edges is proportional to the square root of the number of co-publications; dashed lines indicate a single copublication.
## pubs <- program_publications(foas)
## pubs |> count(pmid) # pmid 32554617 is highly collaborative
copub_data <-
copublication_data(foas, exclude = "32554617") |>
filter(!(
(core_project_num.x %in% excluded_core_project_num) |
(core_project_num.y %in% excluded_core_project_num)
))
nodes <-
copub_data |>
## exclude
tidyr::pivot_longer(dplyr::starts_with("core_project_num")) |>
distinct(id = value) |>
left_join(
project_summary |>
select(id = "core_project_num", project_title) |>
distinct(),
by = "id"
) |>
mutate(
size = 10,
title = paste0(id, ": ", .data$project_title)
) |>
arrange(id)
edges <-
copub_data |>
mutate(
from = core_project_num.x,
to = core_project_num.y,
width = 3 * sqrt(n),
smooth = FALSE,
dashes = n == 1L
)
network <-
visNetwork(nodes, edges) |>
visLayout(randomSeed = 123) |>
visOptions(highlightNearest = TRUE)The figure below shows publications per year…
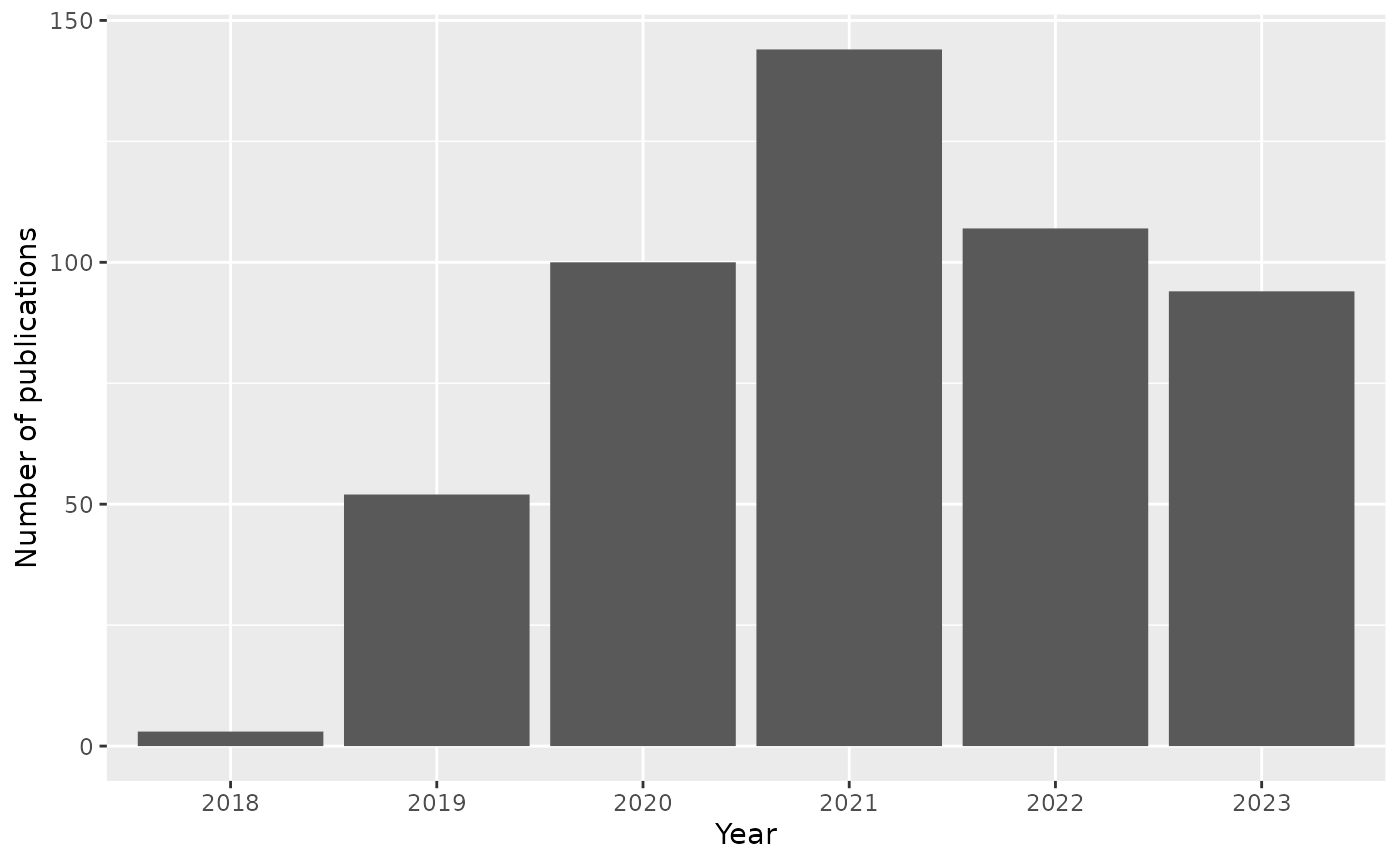
…and the distribution of citations per publication
plot <-
publications |>
filter(citation_count > 0) |>
ggplot(aes(citation_count)) +
scale_x_log10() +
geom_density() +
xlab("Number of citations") + ylab("Number of publications")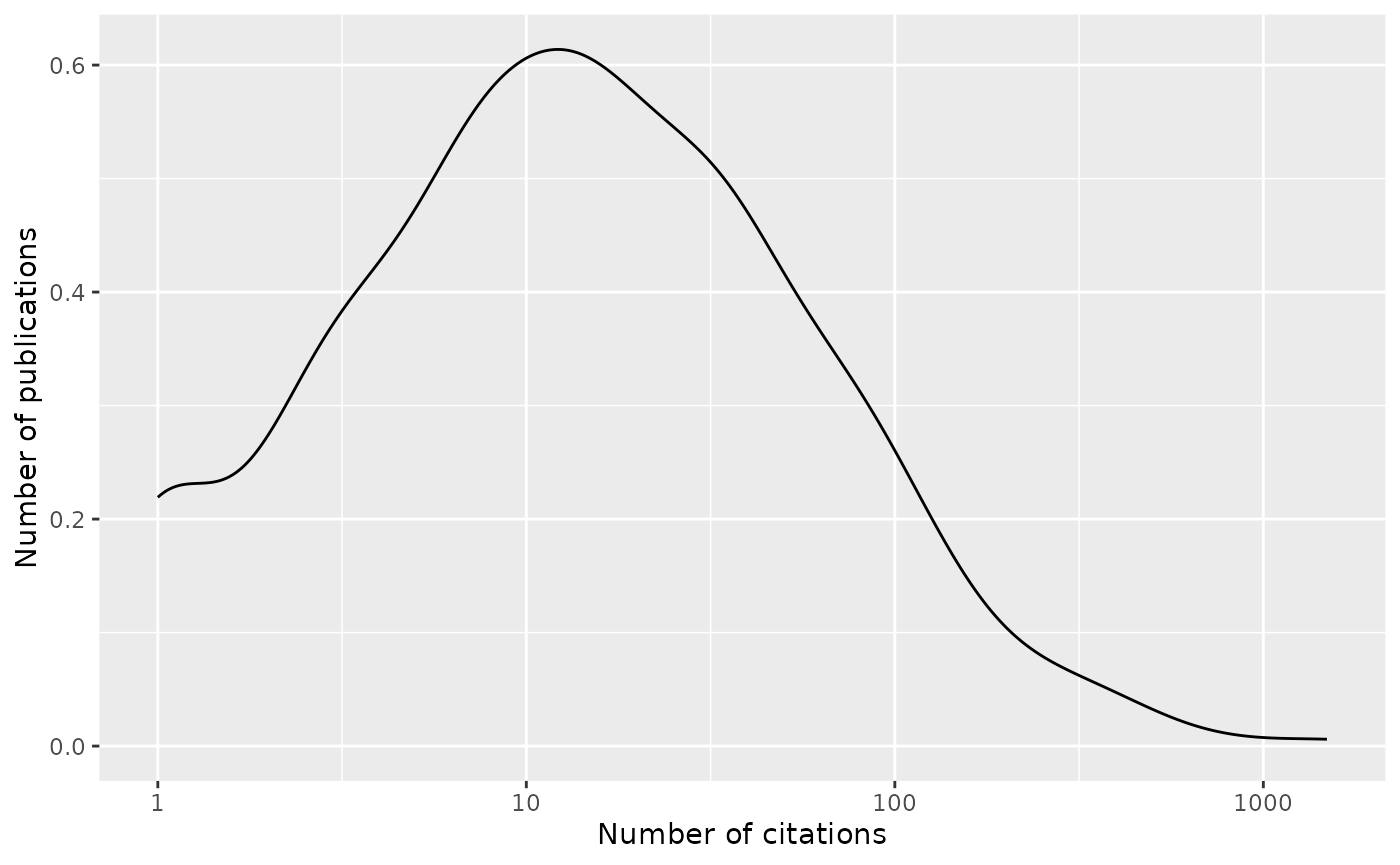
Project-level citations are summarized by the number of publications, the total citation count, and the sum of the ‘relative citation index’, a measure provided by iCite standardizing the impact of publications by year and field of study.
The relationship between amount of funding and relative citation ratio is show in the figure below; mousing over points shows the underlying data and associated core project number. Large ‘U54’ projects do not emphasize publication.
plot <-
citations_by_project |>
ggplot(
aes(
award_amount, total_rcr,
text = paste0(core_project_num, ": ", gpc_shorten(project_title))
)
) +
geom_point() +
scale_x_continuous(labels = scales::comma) +
xlab("Award amount ($)") +
ylab("Total relative citation ratio")Session information
sessionInfo()
#> R version 4.3.2 (2023-10-31)
#> Platform: x86_64-pc-linux-gnu (64-bit)
#> Running under: Ubuntu 22.04.3 LTS
#>
#> Matrix products: default
#> BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
#> LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.20.so; LAPACK version 3.10.0
#>
#> locale:
#> [1] LC_CTYPE=C.UTF-8 LC_NUMERIC=C LC_TIME=C.UTF-8
#> [4] LC_COLLATE=C.UTF-8 LC_MONETARY=C.UTF-8 LC_MESSAGES=C.UTF-8
#> [7] LC_PAPER=C.UTF-8 LC_NAME=C LC_ADDRESS=C
#> [10] LC_TELEPHONE=C LC_MEASUREMENT=C.UTF-8 LC_IDENTIFICATION=C
#>
#> time zone: UTC
#> tzcode source: system (glibc)
#>
#> attached base packages:
#> [1] stats graphics grDevices utils datasets methods base
#>
#> other attached packages:
#> [1] visNetwork_2.1.2 DT_0.31 ggplot2_3.4.4 grantpubcite_0.0.3
#> [5] dplyr_1.1.4
#>
#> loaded via a namespace (and not attached):
#> [1] gtable_0.3.4 xfun_0.41 bslib_0.6.1 htmlwidgets_1.6.4
#> [5] tzdb_0.4.0 rjsoncons_1.0.1 vctrs_0.6.5 tools_4.3.2
#> [9] crosstalk_1.2.1 generics_0.1.3 curl_5.2.0 parallel_4.3.2
#> [13] tibble_3.2.1 fansi_1.0.6 highr_0.10 pkgconfig_2.0.3
#> [17] data.table_1.14.10 desc_1.4.3 lifecycle_1.0.4 compiler_4.3.2
#> [21] farver_2.1.1 stringr_1.5.1 textshaping_0.3.7 munsell_0.5.0
#> [25] htmltools_0.5.7 sass_0.4.8 lazyeval_0.2.2 yaml_2.3.7
#> [29] plotly_4.10.3 pillar_1.9.0 pkgdown_2.0.7 crayon_1.5.2
#> [33] jquerylib_0.1.4 tidyr_1.3.0 ellipsis_0.3.2 cachem_1.0.8
#> [37] tidyselect_1.2.0 digest_0.6.33 stringi_1.8.2 purrr_1.0.2
#> [41] labeling_0.4.3 fastmap_1.1.1 grid_4.3.2 colorspace_2.1-0
#> [45] cli_3.6.1 magrittr_2.0.3 utf8_1.2.4 readr_2.1.4
#> [49] withr_2.5.2 scales_1.3.0 bit64_4.0.5 rmarkdown_2.25
#> [53] httr_1.4.7 bit_4.0.5 ragg_1.2.6 hms_1.1.3
#> [57] memoise_2.0.1 evaluate_0.23 knitr_1.45 viridisLite_0.4.2
#> [61] rlang_1.1.2 glue_1.6.2 vroom_1.6.5 jsonlite_1.8.8
#> [65] R6_2.5.1 systemfonts_1.0.5 fs_1.6.3