Compiled: 2023-12-12
Summary
Note that this is way outside my area of expertise, so I have undoubtedly made terrible blunders.
The ‘grantpubcite’ package can be used to query the NIH Reporter database for funded grants, and the publications associated with those grants. The citation history of publications can be discovered using iCite.
As a case study, suppose we are interested in grants funded under the Information Technology in Cancer Research (ITCR) program.
The Project information section shows that I found 146 projects (a useful sanity check?). The first figure shows that projects funded for more fiscal years received more funding (not too surprising!). At this stage and subsequently I found it useful, using the datatables on the web page, to search for ‘my’ project (U24CA180996, Cancer Genomics: Integrative and Scalable Solutions in R/Bioconductor) and compare how we were doing relative to other projects. Frankly, I did not know where we stood; now I do.
I then focused on projects receiving funding in 2020.
The Maturing projects? subsection uses project title and contact PI to ask whether projects graduate between funding activities. One (Campbell) has matured from ‘Innovative’ to ‘Early-Stage’, three (Griffith, Karchin, and Liu) have matured from ‘Early-stage’ to ‘Advanced’, and two have matured from ‘Advanced’ to ‘Sustained’. I was a little disappointed that there were not more smaller projects maturing.
In the Publications section, we see a positive relationship between funding (amount or duration) and publication. Again this is not surprising, but speaks to the notion that scientific software projects are playing by the same rules as ‘wet lab’ projects; it would be interesting to identify a comparable wet-lab program for comparison. There are some outliers, including one project with a surprisingly large number of publications for the period of the grant, as well as projects with only a handful of publications even after multiple years of funding. The ITCR collaboration section shows that 15 projects collaborated closely enough with each other to be acknowledged in the same publication.
The figure in the Citations section shows what I imagine is a pretty typical pattern, with most publications having moderate (<100) citations, and a few having many citations. Scanning the datatable immediately below the figure indicates that the highly cited publications are biologically driven, where the paper authors have presumably cited the relevant software; the three most-cited publications are all from U24CA180922, which funds the Trinity software. When I look at the five most highly cited of ‘my’ project publications (U24CA180996), they are more modestly successful (181 to 337 citations); interestingly, 4 of the 5 publications emphasize software per se, rather than strictly biological insight.
The ITCR collaborations subsection takes a second look at collaboration, in the sense of ITCR projects citing the works of one another. 38 projects cite work of 30 other projects.
Getting started
See the Introduction to ‘grantpubcite’ article for installation, basic use, and a brief introduction to ‘tidyverse’ operations.
Load the library and other packages to be used in this article.
library(grantpubcite)
library(ggplot2) # also plotly::ggplotly()
library(DT)
library(visNetwork)This document is written in Rmarkdown; code chunks used to generate each table or figure can be shown by toggling the ‘Details’ widget.
Project information
The relevant funding opportunity announcements (FOA) are as follows.
foas <- case_study_foa_itcr()The NIH Reporter contains quite a bit of information about each
grant. Following the Get started
article, we query the reporter_projects() endpoint,
restricting fields included in the return value to a few of particular
interest. Retrieve these fields for all projects in NIH reporter
associated with the funding announcements of interest. Perform some
minor data cleaning by removing leading and trailing whitespace from
contact PI names. The result is a tidyverse tibble, and we
use ‘tidy’ semantics to explore the data. Each project returned by NIH
Reporter, is associated with funding over multiple fiscal years and
mechanisms (e.g., through administrative supplements). A few projects
changed names within the same award; these have been standardized to the
most-recent name.
projects <- program_projects(foas, by = "project")The 161 funded projects are (contact_pi is the most
recent)
The number of projects funded by each type of FOA, and the total amount allcoated to date, is

The number of projects funded per fiscal year is
The following summarizes project funding across years
Maturing projects?
A unique aspect of the ITCR FOA structure is that it envisions
projects starting at one FOA and ‘maturing’ to the next FOA, e.g., an
Early-Stage U01 matures to an Advanced U24. It
is not possible to assess this by tracking a project number through full
FOA, because the project number changes with FOA. Are there projects
with the same contact PI that have received awards from different FOA
tags?
tagged_pis <-
projects |>
distinct(opportunity_number, project_title, contact_pi_name) |>
left_join(foas, by = "opportunity_number") |>
select(foa_tag, contact_pi_name) |>
distinct()
maturing_pis <-
tagged_pis |>
count(contact_pi_name, sort = TRUE) |>
filter(n > 1L) |>
select(-n) |>
left_join(
projects |>
distinct(
opportunity_number, core_project_num, contact_pi_name, project_title
),
by = "contact_pi_name",
multiple = "all"
) |>
left_join(foas, by = "opportunity_number") |>
select(contact_pi_name, project_title, foa_tag) |>
arrange(contact_pi_name, project_title, foa_tag)Direct inspection suggests the following transitions:
Publications
Grantees report publications associated with their grants, and this information can be retrieved from NIH reporter. Queries are formulated in a way similar to projects, as described on the NIH Reporter publication search API, a rich set of query criteria can be used, but fields included in the return are strictly limited.
publications <- program_publications(foas)
## citations: remove duplicates due to collaboration
citations <-
publications |>
select(-c("opportunity_number", "core_project_num")) |>
distinct() |>
select(
"pmid", "citation_count", "relative_citation_ratio",
"field_citation_rate", everything()
) |>
arrange(desc(citation_count))137 projects produced 1993 publications.
Projects and their associated publications and citation statistics are as follows; publications acknowledging more than one grant appear for each acknowledgment. with these projects are obtained with
Project publication and funding
The most prolific projects are
The large number of publications reported by projects ‘U01CA239055’ and ‘U01CA248226’ are apparently because the software tools developed by the group were immediately useful in completing projects at the image analysis center. The relationship between publication and funding amount are visualized as follows.
plot <-
publications_by_project |>
ggplot(aes(
award_amount, n_publ,
text = paste0(core_project_num, ": ", gpc_shorten(project_title))
)) +
geom_point() +
geom_smooth(
method = "lm", formula = y ~ x,
inherit.aes = FALSE, aes(award_amount, n_publ)
) +
scale_x_continuous(labels = scales::comma) +
labs(x = "Total funding ($)", y = "Number of publications")ITCR collaboration
ITCR emphasizes collaboration between funded projects. Are there
examples of collaboration at the level of publication, i.e.,
pmid associated with more than one project number?
Which projects are collaborating through shared publication?
collaborative_projects <-
copublication(foas) |>
left_join(
projects |> select(core_project_num, contact_pi_name, project_title),
by = "core_project_num"
) |>
arrange(desc(collab))Relationships are visualized as a network. Hover over nodes to see project number and title. The width of edges is proportional to the square root of the number of co-publications; dashed lines indicate a single copublication. The strongest relationships are ‘self-copublication’ between grants from the same PI or institution.
copub_data <- copublication_data(foas)
nodes <-
copub_data |>
tidyr::pivot_longer(dplyr::starts_with("core_project_num")) |>
distinct(id = value) |>
left_join(
projects |> select(id = "core_project_num", project_title) |> distinct(),
by = "id"
) |>
mutate(
size = 10,
title = paste0(id, ": ", .data$project_title)
)
edges <-
copub_data |>
mutate(
from = core_project_num.x,
to = core_project_num.y,
width = 3 * sqrt(n),
smooth = FALSE,
dashes = n == 1L
)
copub_network <-
visNetwork(nodes, edges) |>
## set random seed for reproducibility
visLayout(randomSeed = 123) |>
visOptions(
selectedBy = list(variable = "id", highlight = TRUE),
highlightNearest = list(enabled = TRUE, algorithm = "all")
)Citations
Publications and their citation statistics are as follows:
The 1993 publications have 137889 total citations; 220 publications have not been cited; not surprisingly uncited publications are recent.
Highly cited publications are in high-impact journals, and emphasize science-related results rather than a software tool per se – the tool has been used in an important study, and the authors of the study have acknowledged the tool.
Citations follow a very familiar pattern, with a few publications cited frequently.
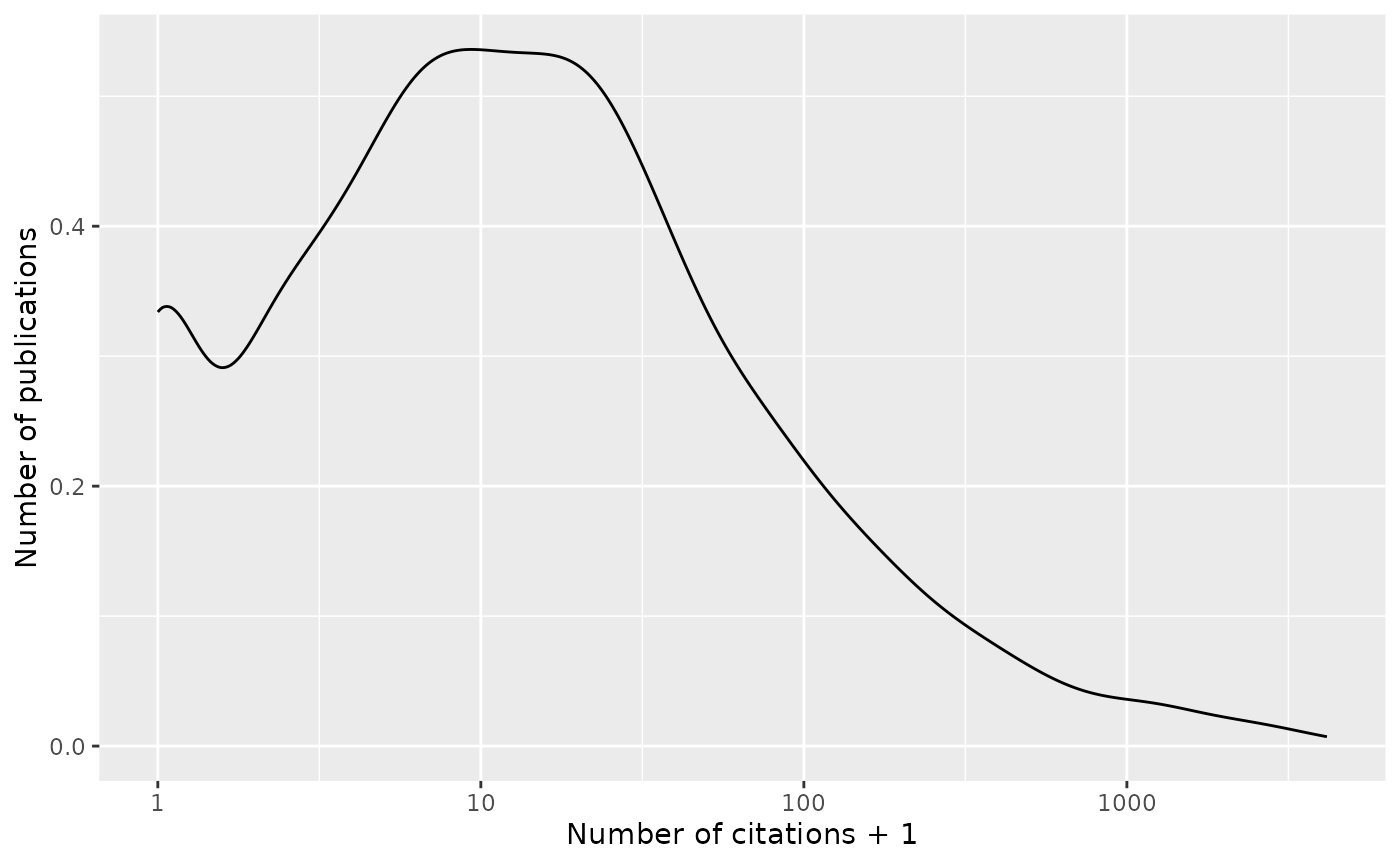
Citations per project are summarized below.
ITCR collaborations
An opportunity for (indirect) collaboration occurs when one project cites the work of another project. Thus we query iCite for the publications that cited ITCR publications, and exclude publications that are not themselves ITCR publications.
cocite_data <- cocitation_data(foas)
## ITCR publications ...
cocite_data |>
distinct(pmid) |>
NROW()
#> [1] 998
## ... cited by other ITCR publications
cocite_data |>
distinct(cited_by) |>
NROW()
#> [1] 1275
## ITCR projects
cocite_data |>
distinct(core_project_num) |>
NROW()
#> [1] 113
## ... cited by other ITCR projects
cocite_data |>
distinct(cited_by_core_project_num) |>
NROW()
#> [1] 125
## pmid / cited_by pairs
total_citations <- NROW(cocite_data)
## self-citations
core_project_num_self_citations <-
cocite_data |>
count(self_citation = core_project_num == cited_by_core_project_num) |>
filter(self_citation) |>
pull(n)There are 998 ITCR publications cited by 1275 ITCR publications. The publications are from 113 ITCR projects cited by 125 ITCR projects.
Of the total 7387 pmid / citation pairs, 2957 are projects citing their own project.
cocitation <-
cocitation(foas) |>
arrange(desc(n_self_citn))Data download
All project information can be downloaded with the following table
Query NIH Reporter and iCite for all publications from all projects, and for extended citation metrics.
Publication ITCR citation data from iCite are
Session information
sessionInfo()
#> R version 4.3.2 (2023-10-31)
#> Platform: x86_64-pc-linux-gnu (64-bit)
#> Running under: Ubuntu 22.04.3 LTS
#>
#> Matrix products: default
#> BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
#> LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.20.so; LAPACK version 3.10.0
#>
#> locale:
#> [1] LC_CTYPE=C.UTF-8 LC_NUMERIC=C LC_TIME=C.UTF-8
#> [4] LC_COLLATE=C.UTF-8 LC_MONETARY=C.UTF-8 LC_MESSAGES=C.UTF-8
#> [7] LC_PAPER=C.UTF-8 LC_NAME=C LC_ADDRESS=C
#> [10] LC_TELEPHONE=C LC_MEASUREMENT=C.UTF-8 LC_IDENTIFICATION=C
#>
#> time zone: UTC
#> tzcode source: system (glibc)
#>
#> attached base packages:
#> [1] stats graphics grDevices utils datasets methods base
#>
#> other attached packages:
#> [1] visNetwork_2.1.2 DT_0.31 ggplot2_3.4.4 grantpubcite_0.0.3
#> [5] dplyr_1.1.4
#>
#> loaded via a namespace (and not attached):
#> [1] gtable_0.3.4 xfun_0.41 bslib_0.6.1 htmlwidgets_1.6.4
#> [5] lattice_0.21-9 tzdb_0.4.0 rjsoncons_1.0.1 vctrs_0.6.5
#> [9] tools_4.3.2 crosstalk_1.2.1 generics_0.1.3 curl_5.2.0
#> [13] parallel_4.3.2 tibble_3.2.1 fansi_1.0.6 highr_0.10
#> [17] pkgconfig_2.0.3 Matrix_1.6-1.1 data.table_1.14.10 desc_1.4.3
#> [21] lifecycle_1.0.4 compiler_4.3.2 farver_2.1.1 stringr_1.5.1
#> [25] textshaping_0.3.7 munsell_0.5.0 htmltools_0.5.7 sass_0.4.8
#> [29] yaml_2.3.7 lazyeval_0.2.2 plotly_4.10.3 tidyr_1.3.0
#> [33] pillar_1.9.0 pkgdown_2.0.7 crayon_1.5.2 jquerylib_0.1.4
#> [37] ellipsis_0.3.2 cachem_1.0.8 nlme_3.1-163 tidyselect_1.2.0
#> [41] digest_0.6.33 stringi_1.8.2 purrr_1.0.2 splines_4.3.2
#> [45] labeling_0.4.3 fastmap_1.1.1 grid_4.3.2 colorspace_2.1-0
#> [49] cli_3.6.1 magrittr_2.0.3 utf8_1.2.4 readr_2.1.4
#> [53] withr_2.5.2 scales_1.3.0 bit64_4.0.5 rmarkdown_2.25
#> [57] httr_1.4.7 bit_4.0.5 ragg_1.2.6 hms_1.1.3
#> [61] memoise_2.0.1 evaluate_0.23 knitr_1.45 viridisLite_0.4.2
#> [65] mgcv_1.9-0 rlang_1.1.2 glue_1.6.2 vroom_1.6.5
#> [69] jsonlite_1.8.8 R6_2.5.1 systemfonts_1.0.5 fs_1.6.3