Introduction & installation
This package is EXPERIMENTAL. It enables access to Globus collection management (including ‘personal’ collections on your own computer), including file and directory transfer. The functions implemented in this package are primarily from the Globus ‘Transfer’ API, documented at https://docs.globus.org/api/transfer/. Many other capabilities of Globus are not implemented.
Install the package if necessary
if (!requireNamespace("remotes", quiety = TRUE))
install.packages("remotes", repos = "https://CRAN.R-project.org")
remotes::install_github("mtmorgan/rglobus")Attach the package to your R session
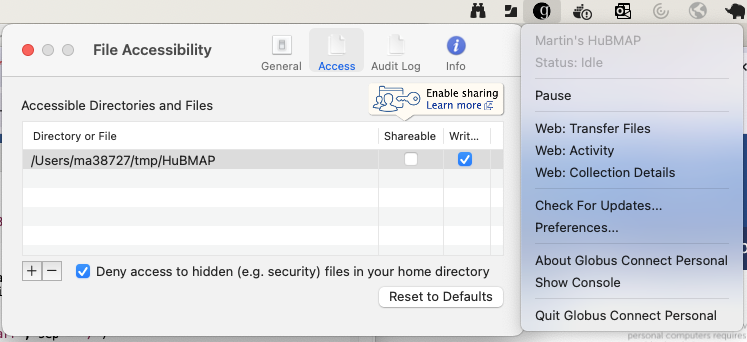
Globus provides software to allow your laptop to appear as a collection. Follow the Globus Connect Personal installation instructions for your operating system, and launch the application. Then identify a location on your local disk to act as a collection.
On macOS, I created a directory ~/tmp/HuBMAP, and then
configured Globus Connect Personal to share that location. I did this by
launching the application (it appears as an icon in the menu bar) then
selecting ‘Preferences’ and using the ‘-’ and ‘+’ buttons to select the
path to my local collection.
Discovering and navigating collections
The functions discussed here are based on the APIs described in Endpoints and Collections and Endpoint and Collection Search.
Globus data sets are organized into collections. Start by discovering collections that contain the words “HuBMAP” and “Public”, in any order.
hubmap_collections <- collections("HuBMAP Public")
hubmap_collections
## # A tibble: 4 × 2
## display_name id
## <chr> <chr>
## 1 HuBMAP Public af603d86-eab9-4eec-bb1d-9d26556741bb
## 2 HuBMAP Dev Public 2b82f085-1d50-4c93-897e-cd79d77481ed
## 3 HuBMAP Stage Public 4b383482-8c5c-48fb-8b80-a450338ca383
## 4 HuBMAP Test Public 1e12c423-dd17-4095-a9f8-12555ee83345Authentication
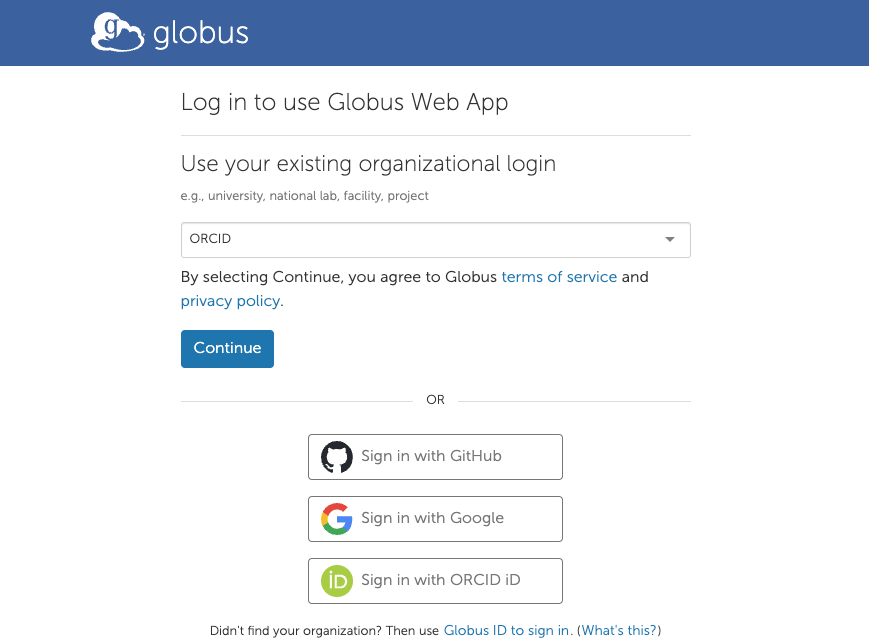
The first time collections() is invoked, Globus requires
that you authenticate. A web page appears, where one can choose
authentication via generic identifiers like ORCID or Google, or through
an institution that you belong to. Follow the browser prompt(s) and
return to R when done.
Collection content
Each collection is presented as directories and files. Focus on the ‘HuBMAP Public’ collection.
hubmap <-
hubmap_collections |>
dplyr::filter(display_name == "HuBMAP Public")List the content of the collection.
globus_ls(hubmap)
## # A tibble: 2,308 × 4
## name last_modified size type
## <chr> <chr> <int> <chr>
## 1 0008a49ac06f4afd886be81491a5a926 2024-07-26 13:35:21+00:00 4096 dir
## 2 0027cb59bcb4a34d5db83acaf934a9d9 2024-07-26 13:19:12+00:00 4096 dir
## 3 002e9747855eef1e69452b39713a7592 2024-08-13 19:04:13+00:00 4096 dir
## 4 00318be0b7cfa3c6ed7fbeab08fe700b 2024-07-26 12:58:42+00:00 4096 dir
## 5 004d4f157df4ba07356cd805131dfc04 2024-08-13 19:21:04+00:00 4096 dir
## 6 0066713ca95c03c52cb40f90ce8bbdb8 2024-04-17 23:24:41+00:00 4096 dir
## 7 007ae59344e7df0e398204ee40155cb0 2024-04-17 23:18:56+00:00 4096 dir
## 8 007f3dfaaa287d5c7c227651f61a9c5b 2024-07-26 14:46:55+00:00 4096 dir
## 9 00cc71c7e1cddac60e794044079faeee 2024-07-26 14:17:08+00:00 4096 dir
## 10 00d1a3623dac388773bc7780fcb42797 2024-04-17 23:02:27+00:00 4096 dir
## # ℹ 2,298 more rowsThere are 2308 records, each corresponding to a HuBMAP dataset.
Information about each dataset, e.g., the dataset with name
0008a49ac06f4afd886be81491a5a926 is available from the HuBMAP
data portal, or using the HuBMAPR package.
(Usually, one would ‘discover’ the dataset name using the data portal or
HuBMAPR, and
then use rglobus to further explore the content).
List the content of a dataset of interest by adding a path to
globus_ls(), e.g.,
path <- "0008a49ac06f4afd886be81491a5a926"
globus_ls(hubmap, path)
## # A tibble: 13 × 4
## name last_modified size type
## <chr> <chr> <int> <chr>
## 1 anndata-zarr 2021-11-21 09:40:50+00:00 4096 dir
## 2 for-visualization 2021-11-21 09:41:41+00:00 4096 dir
## 3 n5 2021-11-21 09:35:26+00:00 4096 dir
## 4 ometiff-pyramids 2021-11-21 09:35:26+00:00 4096 dir
## 5 output_json 2021-11-21 09:39:13+00:00 4096 dir
## 6 output_offsets 2021-11-21 09:37:00+00:00 4096 dir
## 7 sprm_outputs 2021-11-21 09:15:36+00:00 12288 dir
## 8 stitched 2021-11-21 04:44:53+00:00 4096 dir
## 9 experiment.yaml 2021-11-21 04:32:41+00:00 1721 file
## 10 metadata.json 2024-07-26 13:35:21+00:00 90481 file
## 11 pipelineConfig.json 2021-11-21 04:58:21+00:00 5422 file
## 12 session.log 2021-11-21 09:40:55+00:00 18699854 file
## 13 symlinks.tar 2021-11-21 09:17:12+00:00 133120 fileThe dataset consists of files and directories; further explore the content of individual directories by constructing the appropriate path.
path <- paste(path, "anndata-zarr", sep = "/")
globus_ls(hubmap, path)
## # A tibble: 1 × 4
## name last_modified size type
## <chr> <chr> <int> <chr>
## 1 reg1_stitched_expressions-anndata.zarr 2021-11-21 09:40:52+00:00 4096 dir
path <- paste(path, "reg1_stitched_expressions-anndata.zarr", sep = "/")
hubmap |> globus_ls(path)
## # A tibble: 5 × 4
## name last_modified size type
## <chr> <chr> <int> <chr>
## 1 X 2021-11-21 09:40:51+00:00 4096 dir
## 2 layers 2021-11-21 09:40:55+00:00 4096 dir
## 3 obs 2021-11-21 09:40:52+00:00 4096 dir
## 4 obsm 2021-11-21 09:40:52+00:00 4096 dir
## 5 var 2021-11-21 09:40:52+00:00 4096 dirNote in the last example that the functions in rglobus
are designed to support ‘piping’.
Local collections
Collections owned by you appear in collections(), but
the convenience function my_collections() provides another
way to access these.
my_collections <- my_collections()
my_collections
## # A tibble: 1 × 2
## display_name id
## <chr> <chr>
## 1 Martin's HuBMAP 714ce2c4-3268-11ef-9629-453c3ae125a5One aspect of the collection is that the path from the root (starting
with /) or relative to the user home directory needs to be
specified.
globus_ls(my_collections, "/Users/ma38727/tmp")
## # A tibble: 1 × 4
## name last_modified size type
## <chr> <chr> <int> <chr>
## 1 HuBMAP 2024-08-14 21:09:38+00:00 96 dir
path <- "tmp"
globus_ls(my_collections, path)
## # A tibble: 1 × 4
## name last_modified size type
## <chr> <chr> <int> <chr>
## 1 HuBMAP 2024-08-14 21:09:38+00:00 96 dir
## nothing here yet...
path <- "tmp/HuBMAP"
globus_ls(my_collections, path)
## # A tibble: 1 × 4
## name last_modified size type
## <chr> <chr> <int> <chr>
## 1 d1dcab2df80590d8cd8770948abaf976 2024-08-14 21:09:38+00:00 96 dirThe local connection is under our ownership, so it is possible to,
e.g., create a directory; mkdir() returns the updated
directory listing of the enclosing folder.
mkdir(my_collections, "tmp/HuBMAP/test")
## # A tibble: 2 × 4
## name last_modified size type
## <chr> <chr> <int> <chr>
## 1 d1dcab2df80590d8cd8770948abaf976 2024-08-14 21:09:38+00:00 96 dir
## 2 test 2024-08-14 21:09:44+00:00 64 dirOf course we could have used our operating system to create the directory in the path of the local collection.
Directory and file transfer
Directory and file transfer are described in the Task Submission API.
Start this section by ensuring we have the HuBMAP and our own collections
hubmap <-
hubmap_collections |>
dplyr::filter(display_name == "HuBMAP Public")
my_collections <- my_collections()We illustrate directory and file transfer on a specific HuBMAP dataset. Here is the dataset and directory content
hubmap_dataset <- "d1dcab2df80590d8cd8770948abaf976"
globus_ls(hubmap, hubmap_dataset)
## # A tibble: 5 × 4
## name last_modified size type
## <chr> <chr> <int> <chr>
## 1 extras 2023-01-12 15:32:24… 4096 dir
## 2 imzML 2023-01-04 17:56:12… 4096 dir
## 3 ometiffs 2023-01-04 17:58:26… 4096 dir
## 4 d1dcab2df80590d8cd8770948abaf976-metadata.tsv 2023-01-12 15:32:21… 1901 file
## 5 metadata.json 2024-04-17 23:24:32… 34814 fileFile transfer
Use copy() to transfer files or directories between
collections. We start with the metadata.json file, and
transfer it to the ‘test’ directory in our local collection. Define the
source and destination paths in the two collections.
source_path <-
paste(hubmap_dataset, "metadata.json", sep = "/")
destination_path <-
paste("tmp/HuBMAP/test", basename(source_path), sep = "/")Globus anticipates that large data transfers may be involved, so the
copy() operation actually submits a task that runs
asynchronously. Globus has confidence in the robustness of their file
transfer, so adopt a ‘fire and forget’ philosophy – the task will
eventually succeed or fail, perhaps overcoming intermittent network or
other issues.
The ... optional arguments to copy() are
the same as the arguments for the lower-level transfer()
function. By default, transfer tasks send email on completion, but we
will check on task progress and respond appropriately. Each task can be
labeled, the default label is provided by
transfer_label().
task <- copy(
hubmap, my_collections, # collections
source_path, destination_path, # paths
notify_on_succeeded = FALSE
)
task |>
dplyr::glimpse()
## Rows: 1
## Columns: 3
## $ submission_id <chr> "88599a67-5a81-11ef-be9c-83cd94efb466"
## $ task_id <chr> "88599a66-5a81-11ef-be9c-83cd94efb466"
## $ code <chr> "Accepted"We anticipate that code is “Accepted”, indicating that
the task is sufficiently well-formatted to be added to the task
queue.
Task management
Use task_status() to check on status.
task_status(task)
## # A tibble: 1 × 5
## task_id type status nice_status label
## <chr> <chr> <chr> <chr> <chr>
## 1 88599a66-5a81-11ef-be9c-83cd94efb466 TRANSFER ACTIVE Queued 2024-08-14 1…The status column changes from ACTIVE to
SUCCEEDED for successful tasks. An active task proceeding
normally has nice_status either Queued or
OK. An active task may be encountering errors, e.g.,
because the local connection is offline (CONNECTION_FAILED)
or paused (GC_PAUSED) or that the source or destination
file exists but the user does not have permission to read or write it
(PERMISSION_DENIED). Perhaps unintuitively, Globus views
these errors as transient (e.g., because the local collection may come
back online) and so continues to try to complete the task. Active tasks
that persist in an error state will eventually fail.
Failed tasks have status FAILED.
Let’s write a simple loop to check on status, allowing the task to run for up to 60 seconds.
now <- Sys.time()
repeat {
status <- task_status(task)$status
complete <- status %in% c("SUCCEEDED", "FAILED")
if (complete || Sys.time() - now > 60)
break
Sys.sleep(5)
}If the task was successful, we should see the file in our local collection.
status
## [1] "SUCCEEDED"
globus_ls(my_collections, "tmp/HuBMAP/test")
## # A tibble: 1 × 4
## name last_modified size type
## <chr> <chr> <int> <chr>
## 1 metadata.json 2024-08-14 21:09:48+00:00 34814 fileIf the task has failed or is still active for
nice_status reasons that are not likely to resolve, the
task can be canceled. Actually, canceling a completed task generates a
useful message without error.
task_cancel(task)
## TaskComplete: The task completed before the cancel request was processed.Directory transfer
Directory transfer is similar. Here we transfer the entire HuBMAP dataset to our local collection.
Specify the source path as the HuBMAP dataset, and the destination path as the dataset id in our local collection.
source_path <- hubmap_dataset
destination_path <- paste("tmp/HuBMAP", hubmap_dataset, sep = "/")Submit the task as before, but add recursive = TRUE
since this is a directory. As the task may take quite a while to
complete, we will not change the default
notify_on_succeeded option. Check on its initial
status.
task <- copy(
hubmap, my_collections, # collections
source_path, destination_path, # paths
recursive = TRUE
)
task_status(task)
## # A tibble: 1 × 5
## task_id type status nice_status label
## <chr> <chr> <chr> <chr> <chr>
## 1 8c415b64-5a81-11ef-be9c-83cd94efb466 TRANSFER ACTIVE Queued 2024-08-14 1…The task is added to a queue, and eventually the entire content of the HuBMAP dataset is transferred.
One can gain additional insight into the progress of the task by
asking for all_fields of the task status. Relevant fields
are the number of bytes transferred and the effective transfer rate. We
use a helper function to format these values in a more intelligibly.
bytes_to_units <-
function(x)
{
## use R's 'object_size' S3 class to pretty-print bytes as MB, etc
x |>
structure(class = "object_size") |>
format(units = "auto")
}
task_status(task, all_fields = TRUE) |>
dplyr::select(
status, nice_status,
bytes_transferred,
effective_bytes_per_second
) |>
dplyr::mutate(
bytes_transferred = bytes_to_units(bytes_transferred),
effective_bytes_per_second =
bytes_to_units(effective_bytes_per_second)
)
## status nice_status bytes_transferred effective_bytes_per_second
## <chr> <chr> <chr> <chr>
## 1 ACTIVE OK 227.8 Mb 486.9 KbThe transfer can take some time, so in the interest of brevity we cancel the task.
task_cancel(task)
## Canceled: The task has been cancelled successfully.Neglecting to set recursive = TRUE results in a
nice_status IS_A_DIRECTORY. Globus nonetheless
continues to try the file transfer, but we would recognize this as ‘user
error’ and would cancel and resubmit the task.
task <- copy(
hubmap, my_collections, # collections
source_path,
destination_path
)
## ...
task_status(task)
## # A tibble: 1 × 5
## task_id type status nice_status label
## <chr> <chr> <chr> <chr> <chr>
## 1 36bee1e6-5a5b-11ef-be9a-83cd94efb466 TRANSFER ACTIVE IS_A_DIRECTORY 2024-08-1…
## Oops, forgot the `recursive = TRUE` option
task_cancel(task)
task <- copy(
hubmap, my_collections, # collections
source_path,
destination_path,
recursive = TRUE
)Session information
This vignette was compiled using the following software versions
sessionInfo()
## R version 4.4.1 Patched (2024-06-20 r86819)
## Platform: aarch64-apple-darwin23.5.0
## Running under: macOS Sonoma 14.5
##
## Matrix products: default
## BLAS: /Users/ma38727/bin/R-4-4-branch/lib/libRblas.dylib
## LAPACK: /Users/ma38727/bin/R-4-4-branch/lib/libRlapack.dylib; LAPACK version 3.12.0
##
## locale:
## [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
##
## time zone: America/New_York
## tzcode source: internal
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] rglobus_0.0.1
##
## loaded via a namespace (and not attached):
## [1] jsonlite_1.8.8 dplyr_1.1.4 compiler_4.4.1 promises_1.3.0
## [5] Rcpp_1.0.13 tidyselect_1.2.1 later_1.3.2 jquerylib_0.1.4
## [9] systemfonts_1.1.0 textshaping_0.4.0 yaml_2.3.10 fastmap_1.2.0
## [13] R6_2.5.1 rjsoncons_1.3.1 generics_0.1.3 curl_5.2.1
## [17] httr2_1.0.2 knitr_1.48 htmlwidgets_1.6.4 tibble_3.2.1
## [21] desc_1.4.3 openssl_2.2.0 bslib_0.7.0 pillar_1.9.0
## [25] rlang_1.1.4 utf8_1.2.4 cachem_1.1.0 httpuv_1.6.15
## [29] xfun_0.46 fs_1.6.4 sass_0.4.9 cli_3.6.3
## [33] withr_3.0.1 pkgdown_2.1.0 magrittr_2.0.3 digest_0.6.36
## [37] askpass_1.2.0 rappdirs_0.3.3 lifecycle_1.0.4 vctrs_0.6.5
## [41] evaluate_0.24.0 glue_1.7.0 whisker_0.4.1 ragg_1.3.2
## [45] fansi_1.0.6 rmarkdown_2.27 tools_4.4.1 pkgconfig_2.0.3
## [49] htmltools_0.5.8.1