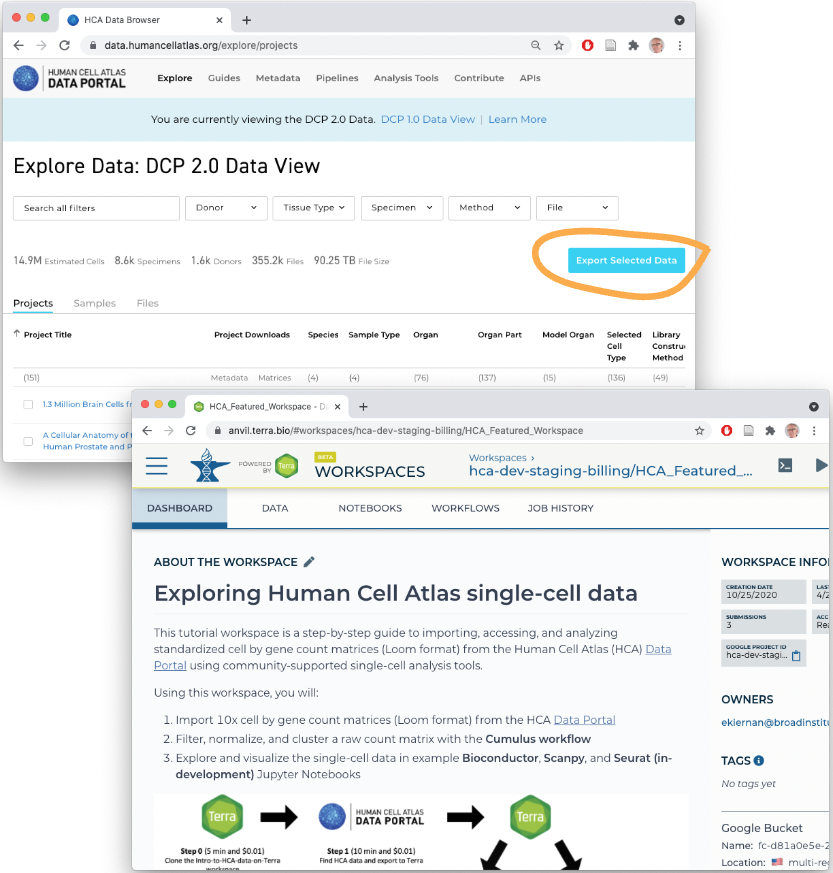
3. Accessing Human Cell Atlas Data on the AnVIL Cloud
Martin Morgan1, Kayla Interdonato2, Yubo Cheng3.
Source:vignettes/C_AnVIL.Rmd
C_AnVIL.RmdOverview
R / Bioconductor packages used
The focus of this vignette is use of data in the AnVIL computational cloud. The AnVIL package provides many functions that facilitate efficient use of this resource.
Time outline
| Activity | Time |
|---|---|
| Introduction to the AnVIL project | 2m |
| AnVIL and the HCA | 2m |
| AnVIL and Bioconductor – the AnVIL package | 3m |
Workshop
Workflows for data reduction – from fastq to loom
Overall workflow
- Researchers design experiments and submit samples to genomics core facilities
- Core facilities sequence samples and generate ‘fastq’ files
- Fastq files are transformed to count matrices summarizing expression of genes in individual cells
- (Perhaps) Core facility performs initial analysis of count matrices
- (Probably) Researcher wishes they could do different variations of the analysis performed by the core facility, especially ‘after’ obtaining the summary count matrix.
What about that fastq-to-count-matrix step?
Many possible routes through this step
Can be formalized, e.g., as ‘Workflow Desccription Language’ (WDL) workflows
-
Example: The HCA Optimus Workflow and detailed documentation
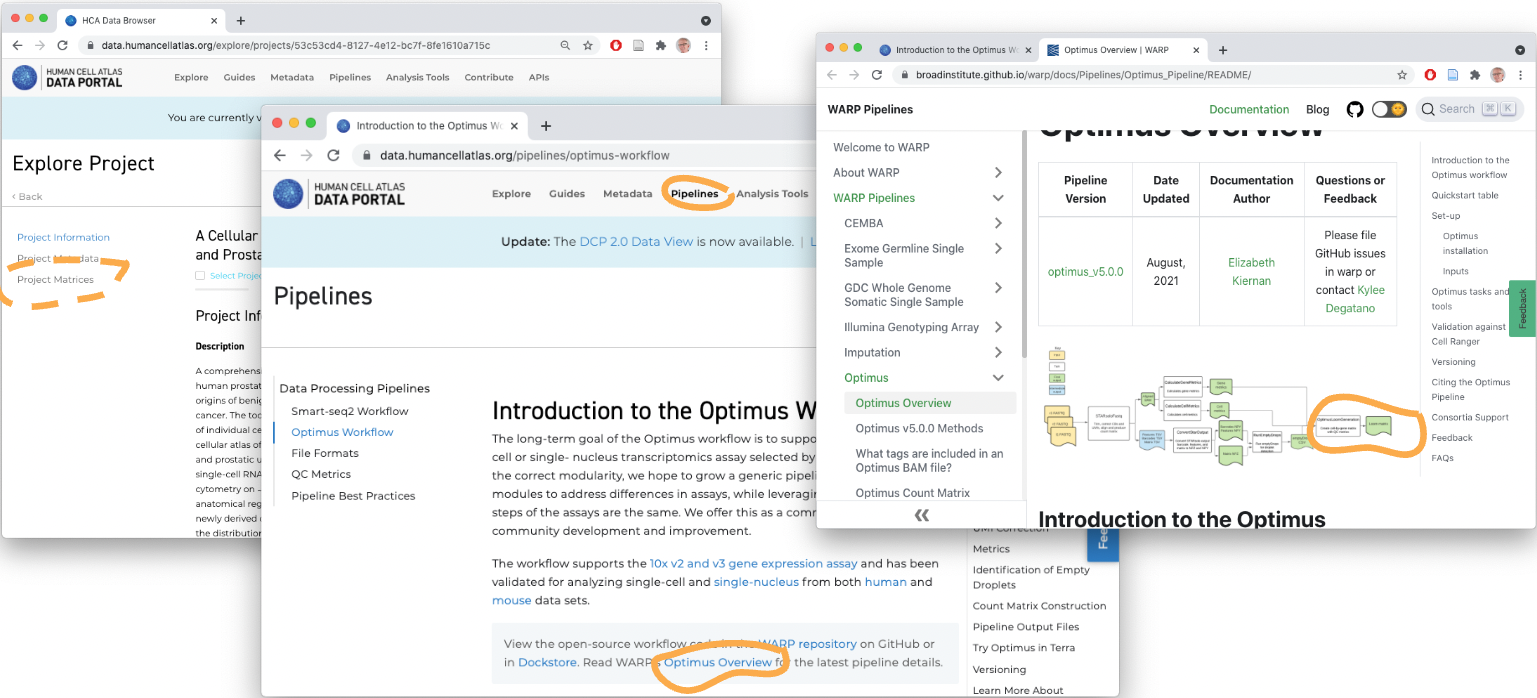
The AnVIL cloud allows this step to be performed repoducibly (reproducibility is not so important as knowing the provenance of the workflow that a formal description provides) and applied to new data sets.
HCA workflows in AnVIL
The count matrix (loom file) is just the first step…
- Many interesting biological questions remain
- These are not really the domain of the core facility focusing on common steps in the analysis of scRNASeq data, but of the individual researcher trying to understand specific biological questions
Importing loom files into AnVIL R / Bioconductor
A standard RStudio environment
- Running on an easily configured container
- E.g., Easy to reconfigure for more memory or disk space
Workflow results deposited in ‘buckets’
- Localize to the local persistent ‘disk’
- Access on the disk as usual, e.g.,
LoomExperiment::import()
Further analysis and visualization withing R / Bioconductor
Introducing OSCA
Installing packages
- Really fast and easy on the AnVIL cloud!
- Container has been pre-configured so system dependencies are satisfied
- Packages are installed as ‘binaries’, so no compiling from source
- Transfer between the binary repository and the RStudio runtime is ‘in the cloud’, so very fast
‘Interactive’ work flows
Acknowledgements
sessionInfo()
#> R version 4.2.0 (2022-04-22)
#> Platform: x86_64-pc-linux-gnu (64-bit)
#> Running under: Ubuntu 20.04.4 LTS
#>
#> Matrix products: default
#> BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
#> LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/liblapack.so.3
#>
#> locale:
#> [1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
#> [3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
#> [5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
#> [7] LC_PAPER=en_US.UTF-8 LC_NAME=C
#> [9] LC_ADDRESS=C LC_TELEPHONE=C
#> [11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
#>
#> attached base packages:
#> [1] stats graphics grDevices utils datasets methods base
#>
#> loaded via a namespace (and not attached):
#> [1] rprojroot_2.0.3 digest_0.6.29 R6_2.5.1 jsonlite_1.8.0
#> [5] magrittr_2.0.3 evaluate_0.15 stringi_1.7.8 rlang_1.0.4
#> [9] cachem_1.0.6 cli_3.3.0 fs_1.5.2 jquerylib_0.1.4
#> [13] bslib_0.4.0 ragg_1.2.2 rmarkdown_2.14 pkgdown_2.0.6
#> [17] textshaping_0.3.6 desc_1.4.1 tools_4.2.0 stringr_1.4.0
#> [21] purrr_0.3.4 yaml_2.3.5 xfun_0.31 fastmap_1.1.0
#> [25] compiler_4.2.0 systemfonts_1.0.4 memoise_2.0.1 htmltools_0.5.3
#> [29] knitr_1.39 sass_0.4.2