C. R and Bioconductor for Genomic Analysis
Source:vignettes/c_course_part_2.Rmd
c_course_part_2.RmdTo use this workshop
- Visit https://workshop.bioconductor.org/
- Register or log-in
Already have a workshop? STOP IT
-
Choose ‘Active workshops’ from the ‘User’ dropdown
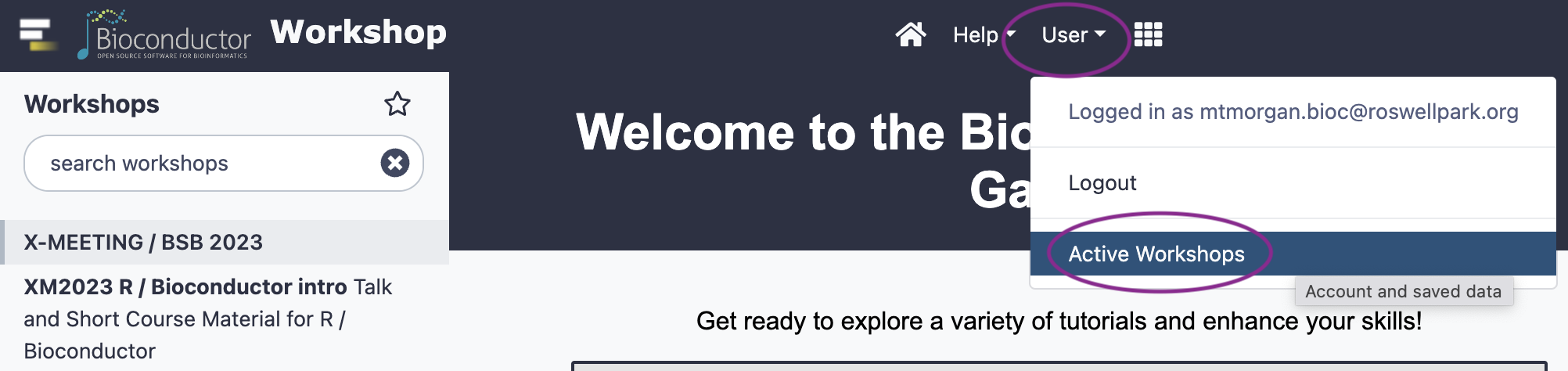
-
Select the existing workshop, and click the ‘Stop’ button
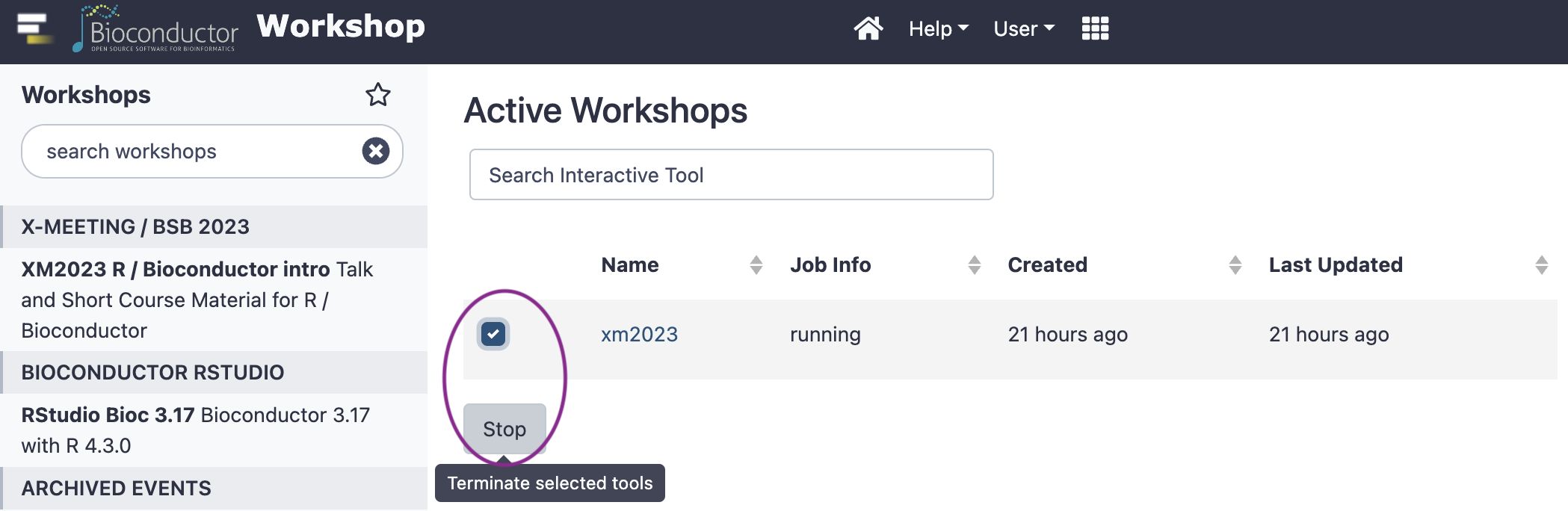
Start a new workshop
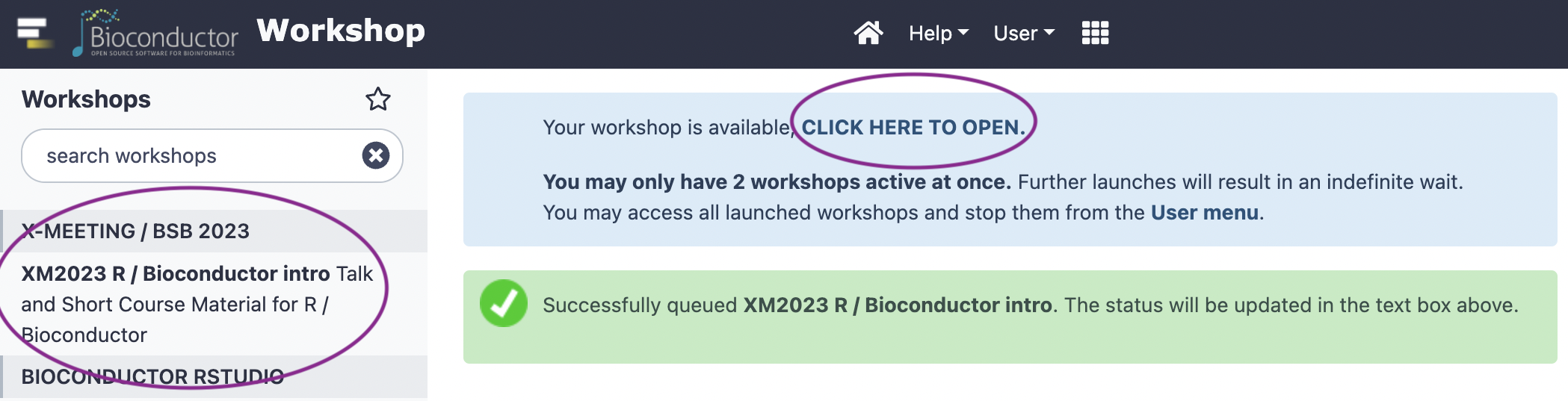
choose the ‘X-MEETING / BBS 2023’
Wait a minute or so
Click to open RStudio
-
In RStudio, choose ‘File’ / ‘Open File…’ / ‘vignettes/c_course_part_2.Rmd’
Introduction
This workshop walks through a single-cellsis with Bioconductor]OSCA (OSCA). The workshop assumes some familiarity with R, and sufficient domain knowledge to know at a superficial level the technical details and scientific motivation for single-cell analysis. The main goal is to increase participants’ confidence in using R to embark on creative and critical data exploration – analysis of single cell data is seldom straight-forward, requiring understanding of methods and critical assessment of data at many steps.
There are four parts to the workshop, probably we will not have time to work through all. The first two provide a high-level overview of how data available in the CELLxGENE portal can be used in the Seurat (Seurat is not a Bioconductor package) and SingleCellExperiment / Bioconductor ecosystem.
The remaining parts walk through two chapters of OSCA, chosen as much because I wanted to learn a bit more about these steps as for any other reason.
Session information
This document was produced with the following R software:
sessionInfo()
#> R version 4.3.0 (2023-04-21)
#> Platform: x86_64-pc-linux-gnu (64-bit)
#> Running under: Ubuntu 22.04.2 LTS
#>
#> Matrix products: default
#> BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
#> LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.20.so; LAPACK version 3.10.0
#>
#> locale:
#> [1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
#> [3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
#> [5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
#> [7] LC_PAPER=en_US.UTF-8 LC_NAME=C
#> [9] LC_ADDRESS=C LC_TELEPHONE=C
#> [11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
#>
#> time zone: Etc/UTC
#> tzcode source: system (glibc)
#>
#> attached base packages:
#> [1] stats graphics grDevices utils datasets methods base
#>
#> loaded via a namespace (and not attached):
#> [1] vctrs_0.6.3 cli_3.6.1 knitr_1.43 rlang_1.1.1
#> [5] xfun_0.39 stringi_1.7.12 purrr_1.0.1 textshaping_0.3.6
#> [9] jsonlite_1.8.5 glue_1.6.2 rprojroot_2.0.3 htmltools_0.5.5
#> [13] ragg_1.2.5 sass_0.4.6 rmarkdown_2.22 evaluate_0.21
#> [17] jquerylib_0.1.4 fastmap_1.1.1 yaml_2.3.7 lifecycle_1.0.3
#> [21] memoise_2.0.1 stringr_1.5.0 compiler_4.3.0 fs_1.6.2
#> [25] systemfonts_1.0.4 digest_0.6.31 R6_2.5.1 magrittr_2.0.3
#> [29] bslib_0.5.0 tools_4.3.0 pkgdown_2.0.7 cachem_1.0.8
#> [33] desc_1.4.2