We use the following packages during this section of the workshop.
## package developed for this workshop
library(XM2023)
## general programming tools
library(dplyr)
## data access
library(cellxgenedp)
## single cell data representation in R
library(SingleCellExperiment) # Bioconductor representation
library(Seurat) # Seurat representationData Transformations
FASTQ files
- DNA sequences and quality scores
- Very large
- Processed e.g., by CellRanger or other software
- Quality control, summary to count matrix (below)
Count matrix
- Usually genes (rows) x cells (columns)
- Can be large (e.g., 30,000 genes x 50,000 cells)
- Usually very sparse, e.g., 95% of cells ‘0’.
- Still large enough to require a decent amount of computing power, e.g., 32 GB RAM, 8 CPU for some steps.
Common formats for count matrix data
- CSV file – lots of zero’s so very wasteful of space.
- ‘Matrix Market’ sparse matrix files, e.g., tuples of <row, column, count> for non-zero values.
- HDF5, e.g., .loom or
.h5ad(anndata). - RDS – An R file, in CELLxGENE these are Seurat objects but in general they could contain any R object.
Matrix representation in R
- In-memory sparse matrices:
dgCMatrixclass from the Matrix package - On-disk representation via Bioconductor’s DelayedArray / HDF5Array.
Counts & annotations
- CSV and Matrix Market files store just counts; usually annotations on columns (e.g., what sample did each cell come from?) are stored separately.
- HDF5 file formats coordinate row and column annotations with count data.
- R software tries to offer a coordinated representation of counts and metadata, e.g., the Seurat or Bioconductor SingleCellExperiment objects.
CELLxGENE
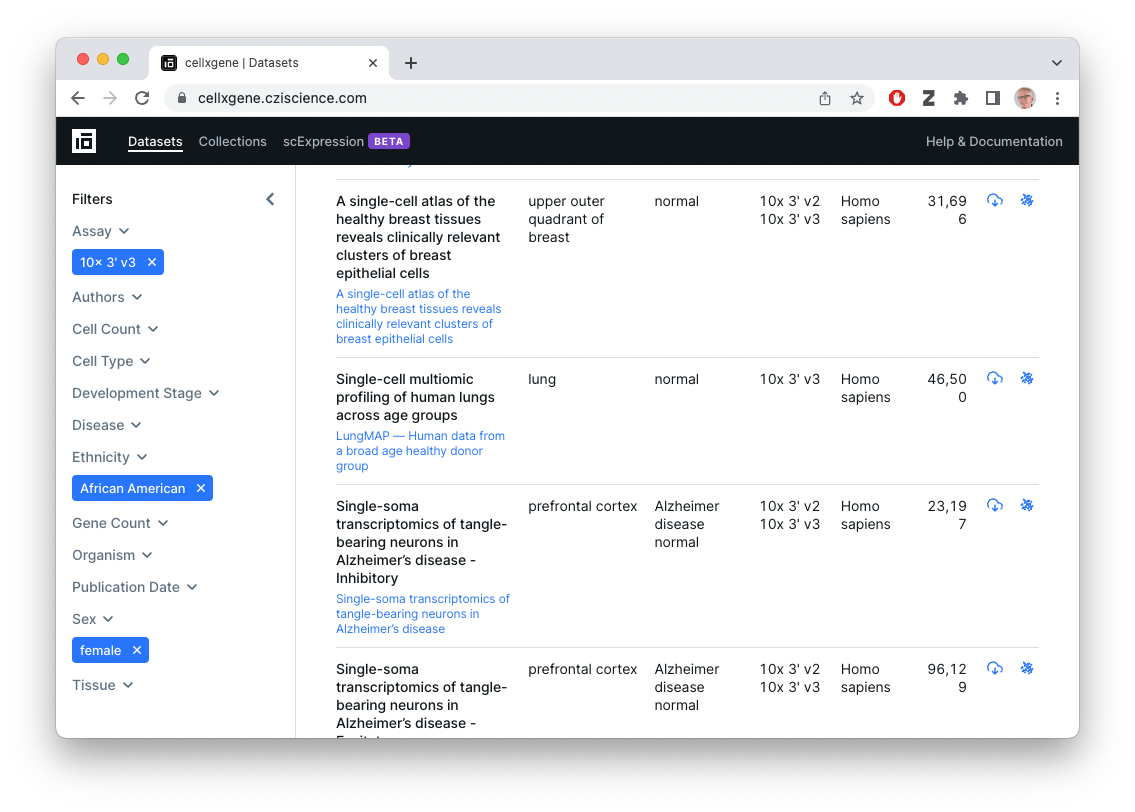
What’s available?
- Collections and datasets contributed by the single-cell community, with some overlap with data sets in the HCA Data Portal.
- FASTQ files
-
.h5ador.RDSsummarized count data and cell metadata, as well as reduced-dimension (e.g., UMAP) representations- Summarized count files provided by the contributor / individual lab, so of uncertain provenance
- Easy to download count data
- Easy to visualize (!)
Programatic Discovery
Why use an R script when the Data Portals exist?
- Easily reproducible
- Flexible exploration of rich & complex data
- Direct integration with Seurat or Bioconductor single-cell workflows
Load the cellxgenedp package
library(cellxgenedp)Retrieve the current database, and use ‘tidy’ functionality to mimic the graphical selection in the web browser – 10x 3’ v3 (EFO:0009922) assay, African American ethnicity, female gender)
db <- db()
african_american_female <-
datasets(db) |>
dplyr::filter(
facets_filter(assay, "ontology_term_id", "EFO:0009922"),
facets_filter(self_reported_ethnicity, "label", "African American"),
facets_filter(sex, "label", "female")
)
african_american_female
#> # A tibble: 32 × 26
#> dataset_id collection_id donor_id assay cell_count cell_type
#> <chr> <chr> <list> <list> <int> <list>
#> 1 8693bc2c-d84f-450f-983a-d… bcb61471-2a4… <list> <list> 304652 <list>
#> 2 37f30005-8a5c-4bdf-88aa-a… bcb61471-2a4… <list> <list> 172847 <list>
#> 3 54914718-c0d3-4e17-9e06-d… bcb61471-2a4… <list> <list> 107344 <list>
#> 4 4c4dedb7-1f74-4be7-9916-2… b953c942-f5d… <list> <list> 23197 <list>
#> 5 8716534a-4c1c-414c-aa98-4… b953c942-f5d… <list> <list> 96129 <list>
#> 6 24205601-0780-4bf2-b1d9-0… c9706a92-0e5… <list> <list> 31696 <list>
#> 7 a963dd1b-ee7e-42b3-8afe-0… a98b828a-622… <list> <list> 48783 <list>
#> 8 36e76662-60b8-4193-8a70-1… 625f6bf4-2f3… <list> <list> 46500 <list>
#> 9 611c3577-81cb-4795-ad3a-4… b9fc3d70-5a7… <list> <list> 49139 <list>
#> 10 0ed93577-0ff5-435e-b599-b… b9fc3d70-5a7… <list> <list> 109995 <list>
#> # ℹ 22 more rows
#> # ℹ 20 more variables: dataset_deployments <chr>, development_stage <list>,
#> # disease <list>, is_primary_data <chr>, is_valid <lgl>,
#> # mean_genes_per_cell <dbl>, name <chr>, organism <list>,
#> # processing_status <list>, published <lgl>, revision <int>,
#> # schema_version <chr>, self_reported_ethnicity <list>, sex <list>,
#> # suspension_type <list>, tissue <list>, tombstone <lgl>, …A particular collection is
collection_id <- "c9706a92-0e5f-46c1-96d8-20e42467f287"
african_american_female |>
dplyr::filter(collection_id %in% .env$collection_id)
#> # A tibble: 1 × 26
#> dataset_id collection_id donor_id assay cell_count cell_type
#> <chr> <chr> <list> <list> <int> <list>
#> 1 24205601-0780-4bf2-b1d9-0e… c9706a92-0e5… <list> <list> 31696 <list>
#> # ℹ 20 more variables: dataset_deployments <chr>, development_stage <list>,
#> # disease <list>, is_primary_data <chr>, is_valid <lgl>,
#> # mean_genes_per_cell <dbl>, name <chr>, organism <list>,
#> # processing_status <list>, published <lgl>, revision <int>,
#> # schema_version <chr>, self_reported_ethnicity <list>, sex <list>,
#> # suspension_type <list>, tissue <list>, tombstone <lgl>, created_at <date>,
#> # published_at <date>, updated_at <date>The dataset associated with this collection is
dataset <-
african_american_female |>
dplyr::filter(collection_id %in% .env$collection_id) |>
dplyr::select(dataset_id)
dataset_id <-
dataset |>
dplyr::pull(dataset_id)
dataset_id
#> [1] "24205601-0780-4bf2-b1d9-0e3cacbc2cd6"The helper function xm2023_cxg_dataset() provides a
summary of this dataset.
xm2023_cxg_dataset(dataset_id)
#> title: A single-cell atlas of the healthy breast tissues reveals
#> clinically relevant clusters of breast epithelial cells
#> description: Single-cell RNA sequencing (scRNA-seq) is an evolving
#> technology used to elucidate the cellular architecture of adult
#> organs. Previous scRNA-seq on breast tissue utilized reduction
#> mammoplasty samples, which are often histologically abnormal. We
#> report a rapid tissue collection/processing protocol to perform
#> scRNA-seq of breast biopsies of healthy women and identify 23
#> breast epithelial cell clusters. Putative cell-of-origin signatures
#> derived from these clusters are applied to analyze transcriptomes
#> of ~3,000 breast cancers. Gene signatures derived from mature
#> luminal cell clusters are enriched in ~68% of breast cancers,
#> whereas a signature from a luminal progenitor cluster is enriched
#> in ~20% of breast cancers. Overexpression of luminal progenitor
#> cluster-derived signatures in HER2+, but not in other subtypes, is
#> associated with unfavorable outcome. We identify TBX3 and PDK4 as
#> genes co-expressed with estrogen receptor (ER) in the normal
#> breasts, and their expression analyses in >550 breast cancers
#> enable prognostically relevant subclassification of ER+ breast
#> cancers.
#> authors: Bhat-Nakshatri, Poornima; Gao, Hongyu; Sheng, Liu; McGuire,
#> Patrick C.; Xuei, Xiaoling; Wan, Jun; Liu, Yunlong; Althouse,
#> Sandra K.; Colter, Austyn; Sandusky, George; Storniolo, Anna Maria;
#> Nakshatri, Harikrishna
#> journal: Cell Reports Medicine
#> assays: 10x 3' v2; 10x 3' v3
#> organism: Homo sapiens
#> ethnicity: African American; Chinese; European‘Join’ selected datasets and files to identify the files associated with these datasets.
selected_files <-
dplyr::left_join(
dataset,
files(db),
by = "dataset_id"
)
selected_files
#> # A tibble: 4 × 8
#> dataset_id file_id filename filetype s3_uri user_submitted created_at
#> <chr> <chr> <chr> <chr> <chr> <lgl> <date>
#> 1 24205601-0780-4bf2… e1c842… "raw.h5… RAW_H5AD s3://… TRUE 1970-01-01
#> 2 24205601-0780-4bf2… 15e9d9… "local.… H5AD s3://… TRUE 1970-01-01
#> 3 24205601-0780-4bf2… 0d3974… "" CXG s3://… TRUE 1970-01-01
#> 4 24205601-0780-4bf2… e254f9… "local.… RDS s3://… TRUE 1970-01-01
#> # ℹ 1 more variable: updated_at <date>Select the first ‘CXG’ file available in this subset of data; for reproducibility we retrieve the dataset id…
selected_files |>
dplyr::filter(filetype == "CXG")
#> # A tibble: 1 × 8
#> dataset_id file_id filename filetype s3_uri user_submitted created_at
#> <chr> <chr> <chr> <chr> <chr> <lgl> <date>
#> 1 24205601-0780-4bf2… 0d3974… "" CXG s3://… TRUE 1970-01-01
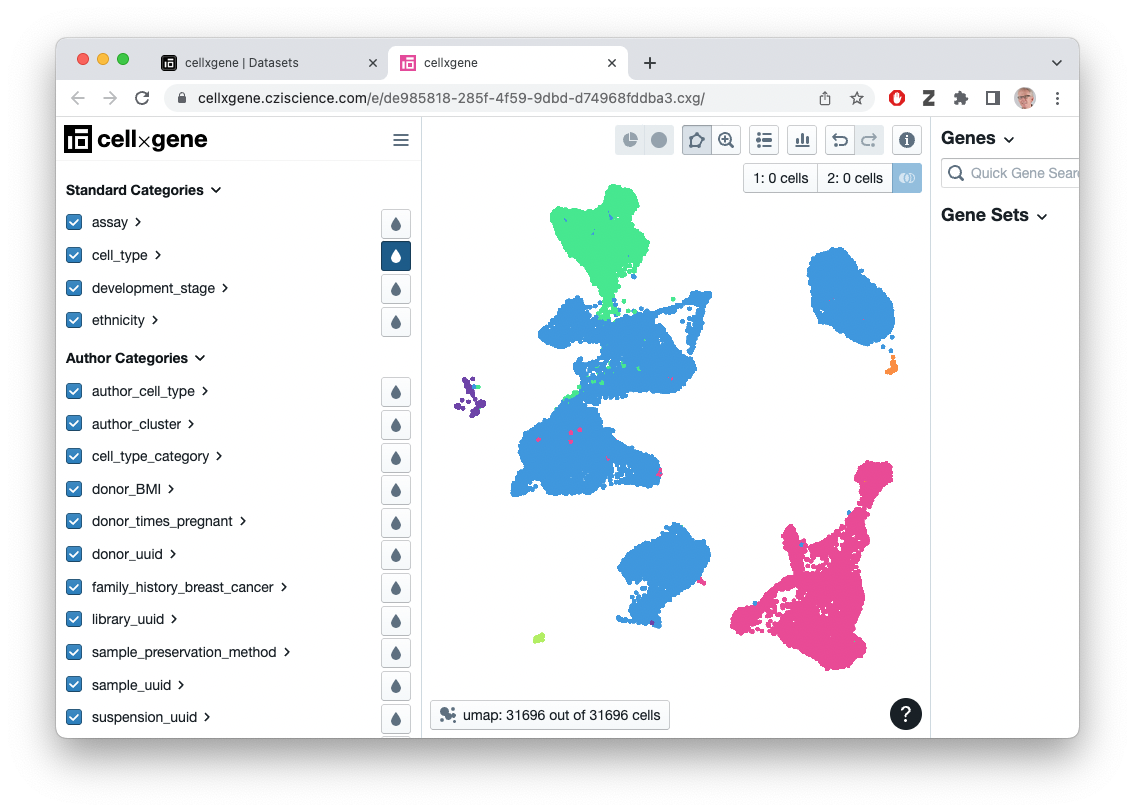
#> # ℹ 1 more variable: updated_at <date>Visualize this ‘CXG’ file in the browser…
selected_files |>
dplyr::filter(filetype == "CXG", dataset_id %in% .env$dataset_id) |>
datasets_visualize()…or select the ‘H5AD’ or ‘RDS’ (Seurat) file associated with the dataset and download it for subsequent processing in R
seurat_file <-
selected_files |>
dplyr::filter(filetype == "RDS", dataset_id %in% .env$dataset_id) |>
files_download(dry.run = FALSE)
h5ad_file <-
selected_files |>
dplyr::filter(filetype == "H5AD", dataset_id == .env$dataset_id) |>
files_download(dry.run = FALSE)The downloaded file is cached, so the next time access is fast.
Seurat
Bioconductor
Data input & representation
The SingleCellExperiment is used to represent
‘rectangular’ single cell expression and other data in R /
Bioconductor. It coordinates a gene x cell count matricies
(assay()) with annotations on the genes
(rowData()) and columns (cellData()), and with
reduced-dimension summaries.
An effective way to represent the .h5ad data as a
SingleCellExperiment is using
zellkonverter::readH5AD().
h5ad <- zellkonverter::readH5AD(h5ad_file, reader = "R", use_hdf5 = TRUE)
#> Registered S3 methods overwritten by 'zellkonverter':
#> method from
#> py_to_r.numpy.ndarray reticulate
#> py_to_r.pandas.core.arrays.categorical.Categorical reticulate
#> Warning in H5Aread(A, ...): Reading attribute data of type 'ENUM' not yet
#> implemented. Values replaced by NA's.
#> Warning in H5Aread(A, ...): Reading attribute data of type 'ENUM' not yet
#> implemented. Values replaced by NA's.
#> Warning in H5Aread(A, ...): Reading attribute data of type 'ENUM' not yet
#> implemented. Values replaced by NA's.
#> Warning in H5Aread(A, ...): Reading attribute data of type 'ENUM' not yet
#> implemented. Values replaced by NA's.
#> Warning in H5Aread(A, ...): Reading attribute data of type 'ENUM' not yet
#> implemented. Values replaced by NA's.
#> Warning in H5Aread(A, ...): Reading attribute data of type 'ENUM' not yet
#> implemented. Values replaced by NA's.
#> Warning in H5Aread(A, ...): Reading attribute data of type 'ENUM' not yet
#> implemented. Values replaced by NA's.
#> Warning in H5Aread(A, ...): Reading attribute data of type 'ENUM' not yet
#> implemented. Values replaced by NA's.
#> Warning in H5Aread(A, ...): Reading attribute data of type 'ENUM' not yet
#> implemented. Values replaced by NA's.
#> Warning in H5Aread(A, ...): Reading attribute data of type 'ENUM' not yet
#> implemented. Values replaced by NA's.
#> Warning in H5Aread(A, ...): Reading attribute data of type 'ENUM' not yet
#> implemented. Values replaced by NA's.
#> Warning in H5Aread(A, ...): Reading attribute data of type 'ENUM' not yet
#> implemented. Values replaced by NA's.
#> Warning in H5Aread(A, ...): Reading attribute data of type 'ENUM' not yet
#> implemented. Values replaced by NA's.
#> Warning in H5Aread(A, ...): Reading attribute data of type 'ENUM' not yet
#> implemented. Values replaced by NA's.
#> Warning in H5Aread(A, ...): Reading attribute data of type 'ENUM' not yet
#> implemented. Values replaced by NA's.
#> Warning in H5Aread(A, ...): Reading attribute data of type 'ENUM' not yet
#> implemented. Values replaced by NA's.
#> Warning in H5Aread(A, ...): Reading attribute data of type 'ENUM' not yet
#> implemented. Values replaced by NA's.
#> Warning in H5Aread(A, ...): Reading attribute data of type 'ENUM' not yet
#> implemented. Values replaced by NA's.
#> Warning in H5Aread(A, ...): Reading attribute data of type 'ENUM' not yet
#> implemented. Values replaced by NA's.
#> Warning in H5Aread(A, ...): Reading attribute data of type 'ENUM' not yet
#> implemented. Values replaced by NA's.
#> Warning in H5Aread(A, ...): Reading attribute data of type 'ENUM' not yet
#> implemented. Values replaced by NA's.
#> Warning in H5Aread(A, ...): Reading attribute data of type 'ENUM' not yet
#> implemented. Values replaced by NA's.
#> Warning in H5Aread(A, ...): Reading attribute data of type 'ENUM' not yet
#> implemented. Values replaced by NA's.
#> Warning in H5Aread(A, ...): Reading attribute data of type 'ENUM' not yet
#> implemented. Values replaced by NA's.
#> Warning in H5Aread(A, ...): Reading attribute data of type 'ENUM' not yet
#> implemented. Values replaced by NA's.
#> Warning in H5Aread(A, ...): Reading attribute data of type 'ENUM' not yet
#> implemented. Values replaced by NA's.
#> Warning in H5Aread(A, ...): Reading attribute data of type 'ENUM' not yet
#> implemented. Values replaced by NA's.
#> Warning in H5Aread(A, ...): Reading attribute data of type 'ENUM' not yet
#> implemented. Values replaced by NA's.
#> Warning in H5Aread(A, ...): Reading attribute data of type 'ENUM' not yet
#> implemented. Values replaced by NA's.
#> Warning in H5Aread(A, ...): Reading attribute data of type 'ENUM' not yet
#> implemented. Values replaced by NA's.
#> Warning in H5Aread(A, ...): Reading attribute data of type 'ENUM' not yet
#> implemented. Values replaced by NA's.
#> Warning in H5Aread(A, ...): Reading attribute data of type 'ENUM' not yet
#> implemented. Values replaced by NA's.Displaying the object…
h5ad
#> class: SingleCellExperiment
#> dim: 33234 31696
#> metadata(3): default_embedding schema_version title
#> assays(1): X
#> rownames(33234): ENSG00000243485 ENSG00000237613 ... ENSG00000277475
#> ENSG00000268674
#> rowData names(4): feature_is_filtered feature_name feature_reference
#> feature_biotype
#> colnames(31696): CMGpool_AAACCCAAGGACAACC CMGpool_AAACCCACAATCTCTT ...
#> K109064_TTTGTTGGTTGCATCA K109064_TTTGTTGGTTGGACCC
#> colData names(34): donor_id self_reported_ethnicity_ontology_term_id
#> ... self_reported_ethnicity development_stage
#> reducedDimNames(3): X_pca X_tsne X_umap
#> mainExpName: NULL
#> altExpNames(0):…suggests the data available and how to access it – there are 33234
genes and 31696 cells. The ‘raw’ data include counts
assays(h5ad, "counts"), annotations on each gene
(rowData()) and cell (colData()), etc…
Working with SingleCellExperiment objects is described
in additional detail subsequent articles.
Session information
This document was produced with the following R software:
sessionInfo()
#> R version 4.3.0 (2023-04-21)
#> Platform: x86_64-pc-linux-gnu (64-bit)
#> Running under: Ubuntu 22.04.2 LTS
#>
#> Matrix products: default
#> BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
#> LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.20.so; LAPACK version 3.10.0
#>
#> locale:
#> [1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
#> [3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
#> [5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
#> [7] LC_PAPER=en_US.UTF-8 LC_NAME=C
#> [9] LC_ADDRESS=C LC_TELEPHONE=C
#> [11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
#>
#> time zone: Etc/UTC
#> tzcode source: system (glibc)
#>
#> attached base packages:
#> [1] stats4 stats graphics grDevices utils datasets methods
#> [8] base
#>
#> other attached packages:
#> [1] SeuratObject_4.1.3 Seurat_4.3.0
#> [3] SingleCellExperiment_1.23.0 SummarizedExperiment_1.31.1
#> [5] Biobase_2.61.0 GenomicRanges_1.53.1
#> [7] GenomeInfoDb_1.37.1 IRanges_2.35.1
#> [9] S4Vectors_0.39.1 BiocGenerics_0.47.0
#> [11] MatrixGenerics_1.13.0 matrixStats_1.0.0
#> [13] cellxgenedp_1.5.0 dplyr_1.1.2
#> [15] XM2023_0.0.1.1
#>
#> loaded via a namespace (and not attached):
#> [1] RColorBrewer_1.1-3 jsonlite_1.8.5 magrittr_2.0.3
#> [4] spatstat.utils_3.0-3 rmarkdown_2.22 fs_1.6.2
#> [7] zlibbioc_1.47.0 ragg_1.2.5 vctrs_0.6.3
#> [10] ROCR_1.0-11 spatstat.explore_3.2-1 memoise_2.0.1
#> [13] RCurl_1.98-1.12 htmltools_0.5.5 S4Arrays_1.1.4
#> [16] curl_5.0.1 Rhdf5lib_1.23.0 rhdf5_2.45.0
#> [19] SparseArray_1.1.10 sctransform_0.3.5 sass_0.4.6
#> [22] parallelly_1.36.0 KernSmooth_2.23-21 bslib_0.5.0
#> [25] basilisk_1.13.1 htmlwidgets_1.6.2 desc_1.4.2
#> [28] ica_1.0-3 plyr_1.8.8 plotly_4.10.2
#> [31] zoo_1.8-12 cachem_1.0.8 igraph_1.5.0
#> [34] mime_0.12 lifecycle_1.0.3 pkgconfig_2.0.3
#> [37] Matrix_1.5-4.1 R6_2.5.1 fastmap_1.1.1
#> [40] GenomeInfoDbData_1.2.10 fitdistrplus_1.1-11 future_1.32.0
#> [43] shiny_1.7.4 digest_0.6.31 colorspace_2.1-0
#> [46] patchwork_1.1.2 tensor_1.5 rprojroot_2.0.3
#> [49] irlba_2.3.5.1 textshaping_0.3.6 filelock_1.0.2
#> [52] progressr_0.13.0 spatstat.sparse_3.0-1 fansi_1.0.4
#> [55] polyclip_1.10-4 abind_1.4-5 httr_1.4.6
#> [58] compiler_4.3.0 withr_2.5.0 rjsoncons_1.0.0
#> [61] HDF5Array_1.29.3 MASS_7.3-60 DelayedArray_0.27.5
#> [64] tools_4.3.0 lmtest_0.9-40 httpuv_1.6.11
#> [67] future.apply_1.11.0 goftest_1.2-3 glue_1.6.2
#> [70] rhdf5filters_1.13.3 nlme_3.1-162 promises_1.2.0.1
#> [73] grid_4.3.0 Rtsne_0.16 reshape2_1.4.4
#> [76] cluster_2.1.4 generics_0.1.3 spatstat.data_3.0-1
#> [79] gtable_0.3.3 tidyr_1.3.0 data.table_1.14.8
#> [82] sp_1.6-1 utf8_1.2.3 XVector_0.41.1
#> [85] spatstat.geom_3.2-1 RcppAnnoy_0.0.20 ggrepel_0.9.3
#> [88] RANN_2.6.1 pillar_1.9.0 stringr_1.5.0
#> [91] later_1.3.1 splines_4.3.0 lattice_0.21-8
#> [94] deldir_1.0-9 survival_3.5-5 tidyselect_1.2.0
#> [97] miniUI_0.1.1.1 pbapply_1.7-0 knitr_1.43
#> [100] gridExtra_2.3 scattermore_1.2 xfun_0.39
#> [103] DT_0.28 stringi_1.7.12 lazyeval_0.2.2
#> [106] yaml_2.3.7 evaluate_0.21 codetools_0.2-19
#> [109] tibble_3.2.1 cli_3.6.1 uwot_0.1.14
#> [112] xtable_1.8-4 reticulate_1.30 systemfonts_1.0.4
#> [115] munsell_0.5.0 jquerylib_0.1.4 zellkonverter_1.11.1
#> [118] Rcpp_1.0.10 dir.expiry_1.9.0 spatstat.random_3.1-5
#> [121] globals_0.16.2 png_0.1-8 parallel_4.3.0
#> [124] ellipsis_0.3.2 pkgdown_2.0.7 ggplot2_3.4.2
#> [127] basilisk.utils_1.13.1 bitops_1.0-7 listenv_0.9.0
#> [130] viridisLite_0.4.2 scales_1.2.1 ggridges_0.5.4
#> [133] leiden_0.4.3 purrr_1.0.1 crayon_1.5.2
#> [136] rlang_1.1.1 cowplot_1.1.1